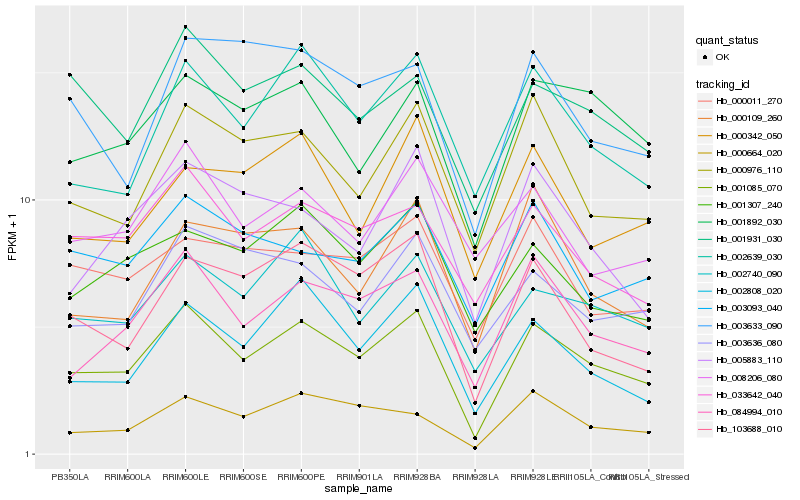
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001085_070 |
0.0 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |
| 2 |
Hb_084994_010 |
0.0999594083 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002740_090 |
0.107829696 |
- |
- |
PREDICTED: putative membrane-bound O-acyltransferase C24H6.01c [Jatropha curcas] |
| 4 |
Hb_002808_020 |
0.1122800414 |
- |
- |
PREDICTED: uncharacterized protein LOC105630318 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001892_030 |
0.1125543004 |
- |
- |
PREDICTED: phytanoyl-CoA dioxygenase isoform X1 [Jatropha curcas] |
| 6 |
Hb_103688_010 |
0.1126579714 |
- |
- |
PREDICTED: sucrose-phosphatase 2-like [Jatropha curcas] |
| 7 |
Hb_002639_030 |
0.1131863914 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 8 |
Hb_000664_020 |
0.1141850052 |
- |
- |
tryptophanyl-tRNA synthetase, putative [Ricinus communis] |
| 9 |
Hb_000976_110 |
0.114912381 |
- |
- |
PREDICTED: KRR1 small subunit processome component homolog [Jatropha curcas] |
| 10 |
Hb_033642_040 |
0.1163683981 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001931_030 |
0.118389042 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta-like [Nelumbo nucifera] |
| 12 |
Hb_001307_240 |
0.1228550212 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003093_040 |
0.1265101983 |
- |
- |
PREDICTED: nucleolar complex protein 4 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_003636_080 |
0.1282152586 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 15 |
Hb_005883_110 |
0.1284142668 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_003633_090 |
0.1289631892 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 17 |
Hb_000011_270 |
0.1292057491 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 18 |
Hb_000109_260 |
0.1309504427 |
- |
- |
PREDICTED: WAT1-related protein At3g28050-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000342_050 |
0.1317740868 |
- |
- |
hypothetical protein POPTR_0006s24710g [Populus trichocarpa] |
| 20 |
Hb_008206_080 |
0.1320791987 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 17 [Jatropha curcas] |