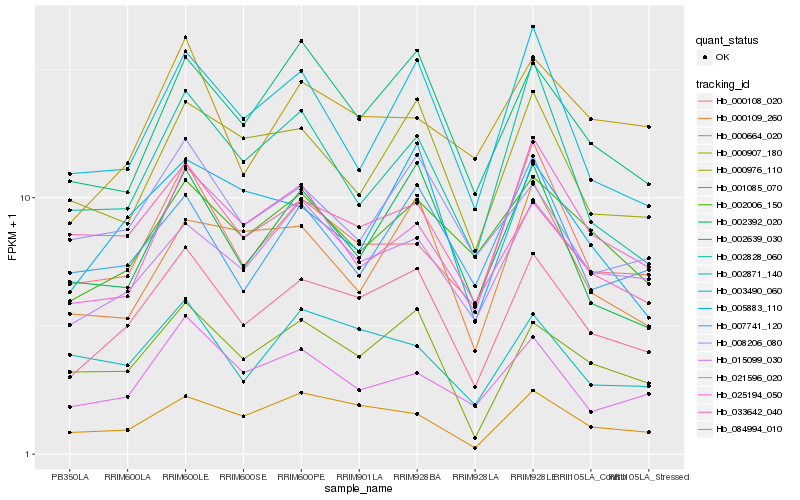
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_084994_010 |
0.0 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_015099_030 |
0.0995054574 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 3 |
Hb_001085_070 |
0.0999594083 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |
| 4 |
Hb_002639_030 |
0.1001523528 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 5 |
Hb_000664_020 |
0.1036687397 |
- |
- |
tryptophanyl-tRNA synthetase, putative [Ricinus communis] |
| 6 |
Hb_003490_060 |
0.109045696 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic isoform X2 [Sesamum indicum] |
| 7 |
Hb_002392_020 |
0.1100249025 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 8 |
Hb_002006_150 |
0.1108226213 |
- |
- |
copine, putative [Ricinus communis] |
| 9 |
Hb_000976_110 |
0.1108941007 |
- |
- |
PREDICTED: KRR1 small subunit processome component homolog [Jatropha curcas] |
| 10 |
Hb_000108_020 |
0.1143286472 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 11 |
Hb_005883_110 |
0.1157328888 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_021596_020 |
0.1176537876 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 13 |
Hb_033642_040 |
0.1195109995 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 14 |
Hb_008206_080 |
0.1223548692 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 17 [Jatropha curcas] |
| 15 |
Hb_000907_180 |
0.1224559371 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002871_140 |
0.1229199801 |
- |
- |
hypothetical protein JCGZ_06847 [Jatropha curcas] |
| 17 |
Hb_002828_060 |
0.1230530857 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000109_260 |
0.1233503913 |
- |
- |
PREDICTED: WAT1-related protein At3g28050-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_025194_050 |
0.1247972135 |
- |
- |
PREDICTED: uncharacterized protein LOC105644851 isoform X1 [Jatropha curcas] |
| 20 |
Hb_007741_120 |
0.125142809 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 4, chloroplastic [Jatropha curcas] |