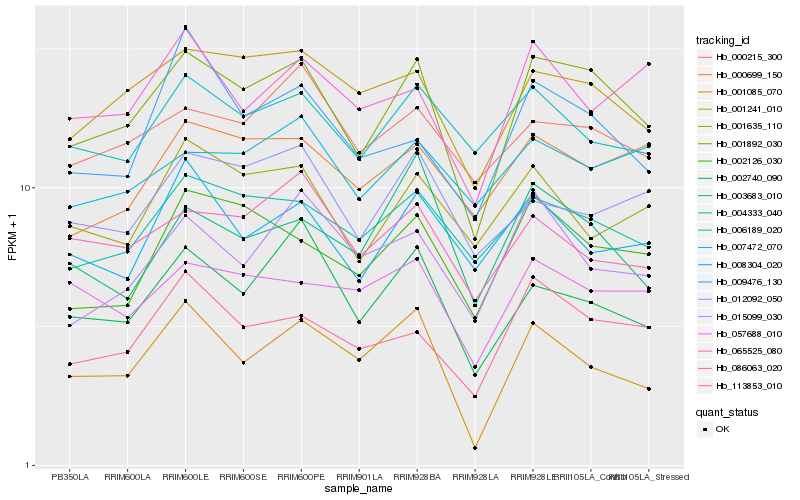
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001892_030 |
0.0 |
- |
- |
PREDICTED: phytanoyl-CoA dioxygenase isoform X1 [Jatropha curcas] |
| 2 |
Hb_001241_010 |
0.0882904318 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_113853_010 |
0.0925372744 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |
| 4 |
Hb_004333_040 |
0.0956039705 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 5 |
Hb_002126_030 |
0.0994950037 |
- |
- |
ferrochelatase, putative [Ricinus communis] |
| 6 |
Hb_002740_090 |
0.0995172787 |
- |
- |
PREDICTED: putative membrane-bound O-acyltransferase C24H6.01c [Jatropha curcas] |
| 7 |
Hb_057688_010 |
0.1044929827 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000215_300 |
0.1063757478 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 9 |
Hb_006189_020 |
0.1085160159 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 10 |
Hb_000699_150 |
0.1086217576 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 11 |
Hb_012092_050 |
0.1101894227 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_086063_020 |
0.111036423 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 13 |
Hb_065525_080 |
0.1112822085 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 14 [Populus euphratica] |
| 14 |
Hb_008304_020 |
0.1121936381 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |
| 15 |
Hb_009476_130 |
0.1124597126 |
- |
- |
hypothetical protein VITISV_005430 [Vitis vinifera] |
| 16 |
Hb_001085_070 |
0.1125543004 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |
| 17 |
Hb_007472_070 |
0.1130335414 |
- |
- |
cir, putative [Ricinus communis] |
| 18 |
Hb_003683_010 |
0.1131881412 |
- |
- |
longevity assurance factor, putative [Ricinus communis] |
| 19 |
Hb_015099_030 |
0.1133326191 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 20 |
Hb_001635_110 |
0.1138497286 |
- |
- |
PREDICTED: NADP-specific glutamate dehydrogenase [Jatropha curcas] |