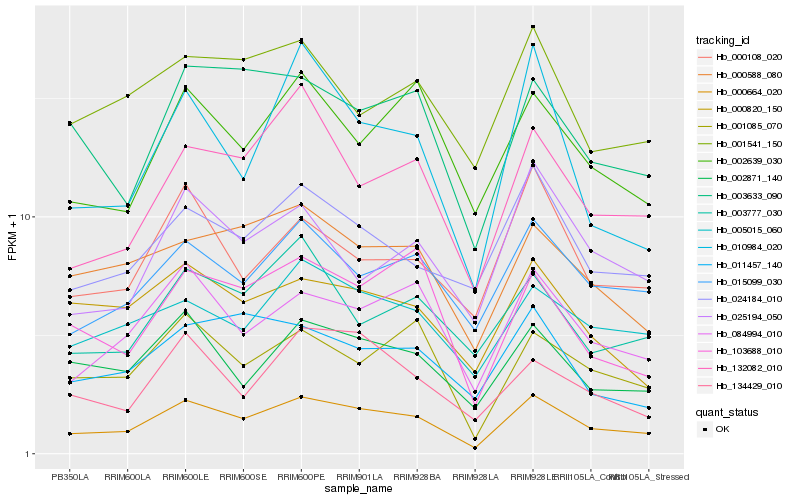
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000664_020 |
0.0 |
- |
- |
tryptophanyl-tRNA synthetase, putative [Ricinus communis] |
| 2 |
Hb_084994_010 |
0.1036687397 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_015099_030 |
0.1105118572 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 4 |
Hb_002871_140 |
0.111635485 |
- |
- |
hypothetical protein JCGZ_06847 [Jatropha curcas] |
| 5 |
Hb_132082_010 |
0.1129319653 |
- |
- |
hypothetical protein VITISV_030895 [Vitis vinifera] |
| 6 |
Hb_001085_070 |
0.1141850052 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |
| 7 |
Hb_024184_010 |
0.1142439907 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 8 |
Hb_005015_060 |
0.1148905537 |
- |
- |
PREDICTED: developmentally-regulated G-protein 3 [Jatropha curcas] |
| 9 |
Hb_000588_080 |
0.1200883385 |
- |
- |
PREDICTED: uncharacterized protein LOC105638459 [Jatropha curcas] |
| 10 |
Hb_010984_020 |
0.1233064974 |
- |
- |
Translation factor GUF1 like, chloroplastic [Glycine soja] |
| 11 |
Hb_003633_090 |
0.123895503 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 12 |
Hb_103688_010 |
0.1238962597 |
- |
- |
PREDICTED: sucrose-phosphatase 2-like [Jatropha curcas] |
| 13 |
Hb_002639_030 |
0.124081904 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 14 |
Hb_000108_020 |
0.1253520745 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 15 |
Hb_134429_010 |
0.1259650267 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000820_150 |
0.1287815501 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9-like [Jatropha curcas] |
| 17 |
Hb_003777_030 |
0.1300777188 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 18 |
Hb_011457_140 |
0.1306826563 |
- |
- |
PREDICTED: uncharacterized protein LOC105649947 [Jatropha curcas] |
| 19 |
Hb_025194_050 |
0.131366372 |
- |
- |
PREDICTED: uncharacterized protein LOC105644851 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001541_150 |
0.1316928226 |
- |
- |
conserved hypothetical protein [Ricinus communis] |