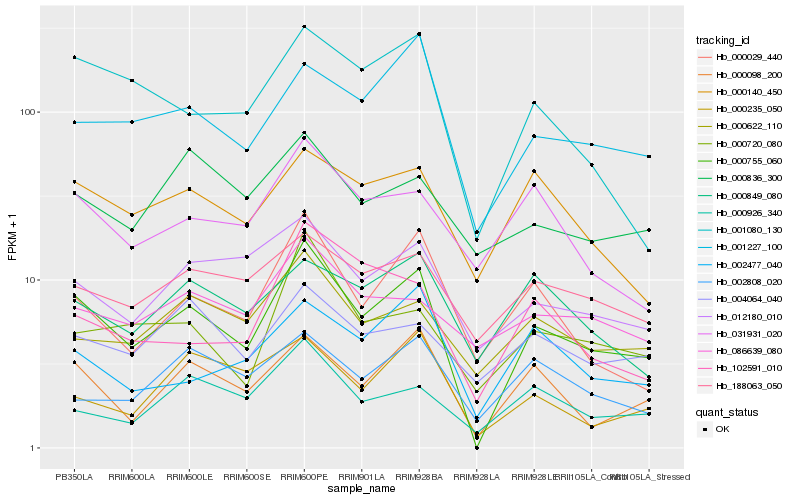
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000755_060 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104424740 [Eucalyptus grandis] |
| 2 |
Hb_000098_200 |
0.1541457754 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7-like isoform X2 [Populus euphratica] |
| 3 |
Hb_001227_100 |
0.1687775504 |
- |
- |
RecName: Full=Enolase 1; AltName: Full=2-phospho-D-glycerate hydro-lyase 1; AltName: Full=2-phosphoglycerate dehydratase 1; AltName: Allergen=Hev b 9 [Hevea brasiliensis] |
| 4 |
Hb_000029_440 |
0.1705766201 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit isoform X2 [Vitis vinifera] |
| 5 |
Hb_001080_130 |
0.1725422495 |
- |
- |
catalase [Hevea brasiliensis] |
| 6 |
Hb_012180_010 |
0.1746697974 |
- |
- |
PREDICTED: (S)-ureidoglycine aminohydrolase [Jatropha curcas] |
| 7 |
Hb_000720_080 |
0.1805972266 |
- |
- |
PREDICTED: bifunctional UDP-glucose 4-epimerase and UDP-xylose 4-epimerase 1 [Jatropha curcas] |
| 8 |
Hb_000622_110 |
0.1808360999 |
- |
- |
cmp-sialic acid transporter, putative [Ricinus communis] |
| 9 |
Hb_000926_340 |
0.1834847634 |
- |
- |
PREDICTED: EH domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000140_450 |
0.1856763707 |
- |
- |
PREDICTED: calreticulin-3-like [Populus euphratica] |
| 11 |
Hb_188063_050 |
0.1872780676 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 12 |
Hb_002477_040 |
0.1872895376 |
- |
- |
P-loop containing nucleoside triphosphate hydrolases superfamily protein isoform 2 [Theobroma cacao] |
| 13 |
Hb_031931_020 |
0.189225218 |
- |
- |
Minor histocompatibility antigen H13, putative [Ricinus communis] |
| 14 |
Hb_000235_050 |
0.1907422451 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_102591_010 |
0.1908553937 |
- |
- |
hypothetical protein JCGZ_09876 [Jatropha curcas] |
| 16 |
Hb_004064_040 |
0.1926014135 |
- |
- |
PREDICTED: Down syndrome critical region protein 3 homolog isoform X3 [Jatropha curcas] |
| 17 |
Hb_000849_080 |
0.1947652476 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKU5 isoform X1 [Jatropha curcas] |
| 18 |
Hb_086639_080 |
0.1951130235 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002808_020 |
0.1969813631 |
- |
- |
PREDICTED: uncharacterized protein LOC105630318 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000836_300 |
0.1972680546 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |