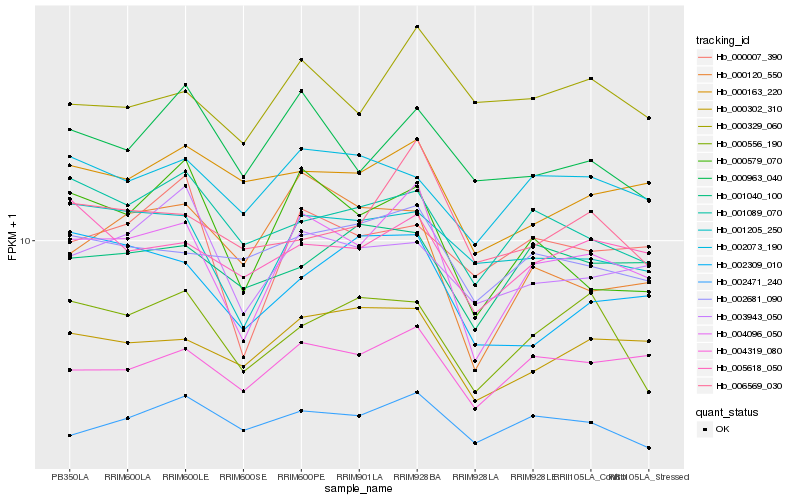
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004096_050 |
0.0 |
- |
- |
hypothetical protein B456_006G229700 [Gossypium raimondii] |
| 2 |
Hb_004319_080 |
0.1030658513 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 1 [Jatropha curcas] |
| 3 |
Hb_000302_310 |
0.1107308806 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 4 |
Hb_000579_070 |
0.1135545176 |
- |
- |
hypothetical protein CISIN_1g0119542mg, partial [Citrus sinensis] |
| 5 |
Hb_001205_250 |
0.1150296821 |
- |
- |
PREDICTED: uncharacterized protein LOC105643856 [Jatropha curcas] |
| 6 |
Hb_005618_050 |
0.122417395 |
- |
- |
PREDICTED: charged multivesicular body protein 7 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002681_090 |
0.1230685664 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000007_390 |
0.1246168311 |
- |
- |
hypothetical protein JCGZ_18250 [Jatropha curcas] |
| 9 |
Hb_006569_030 |
0.1250612059 |
- |
- |
PREDICTED: uncharacterized protein LOC105650907 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002471_240 |
0.1251288858 |
- |
- |
PREDICTED: protein POLLEN DEFECTIVE IN GUIDANCE 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000556_190 |
0.1254953102 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000163_220 |
0.1258663568 |
- |
- |
hypothetical protein CISIN_1g022301mg [Citrus sinensis] |
| 13 |
Hb_000120_550 |
0.1265243116 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 14 |
Hb_001089_070 |
0.1265630579 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 15 |
Hb_002073_190 |
0.1266598475 |
- |
- |
PREDICTED: uncharacterized protein LOC105649812 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003943_050 |
0.1272578226 |
- |
- |
phosphoglucomutase, putative [Ricinus communis] |
| 17 |
Hb_000329_060 |
0.1285823484 |
- |
- |
chloroplast 5-enolpyruvylshikimate 3-phosphate synthase [Hevea brasiliensis] |
| 18 |
Hb_001040_100 |
0.1286935308 |
- |
- |
PREDICTED: general transcription factor IIH subunit 4 [Jatropha curcas] |
| 19 |
Hb_000963_040 |
0.1287212548 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 20 |
Hb_002309_010 |
0.1301425013 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |