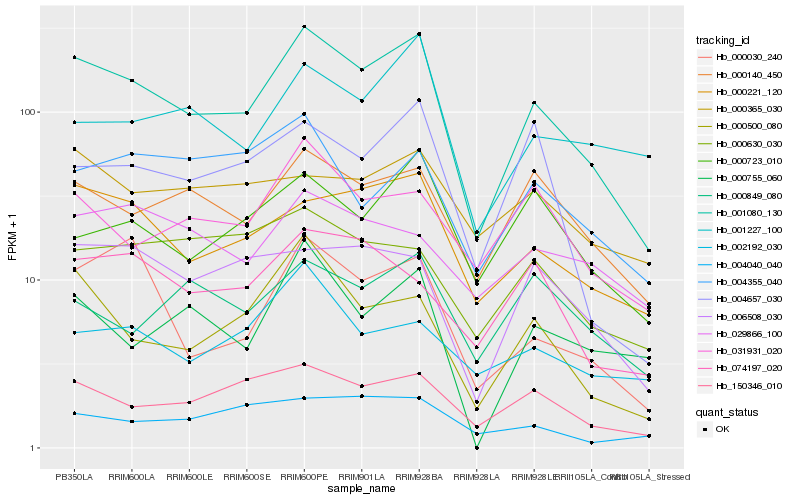
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001080_130 |
0.0 |
- |
- |
catalase [Hevea brasiliensis] |
| 2 |
Hb_000221_120 |
0.1284014991 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 3 |
Hb_150346_010 |
0.1395066768 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 4 |
Hb_000140_450 |
0.1451303128 |
- |
- |
PREDICTED: calreticulin-3-like [Populus euphratica] |
| 5 |
Hb_000500_080 |
0.1504401416 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 6 |
Hb_000030_240 |
0.1548426677 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_006508_030 |
0.1583218844 |
- |
- |
PREDICTED: CDPK-related kinase 1-like [Jatropha curcas] |
| 8 |
Hb_031931_020 |
0.1657531543 |
- |
- |
Minor histocompatibility antigen H13, putative [Ricinus communis] |
| 9 |
Hb_000723_010 |
0.1688581527 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_004657_030 |
0.1693566672 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 11 |
Hb_001227_100 |
0.1703530526 |
- |
- |
RecName: Full=Enolase 1; AltName: Full=2-phospho-D-glycerate hydro-lyase 1; AltName: Full=2-phosphoglycerate dehydratase 1; AltName: Allergen=Hev b 9 [Hevea brasiliensis] |
| 12 |
Hb_074197_020 |
0.1708321708 |
- |
- |
PREDICTED: uncharacterized protein LOC105645948 [Jatropha curcas] |
| 13 |
Hb_000755_060 |
0.1725422495 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104424740 [Eucalyptus grandis] |
| 14 |
Hb_000630_030 |
0.1740490485 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 isoform X1 [Populus euphratica] |
| 15 |
Hb_004355_040 |
0.1754955209 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 16 |
Hb_000365_030 |
0.1790650084 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 17 |
Hb_002192_030 |
0.179500042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000849_080 |
0.1797484343 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKU5 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004040_040 |
0.1803938038 |
desease resistance |
Gene Name: ABC_tran |
hypothetical protein POPTR_0006s07370g [Populus trichocarpa] |
| 20 |
Hb_029866_100 |
0.1818117431 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |