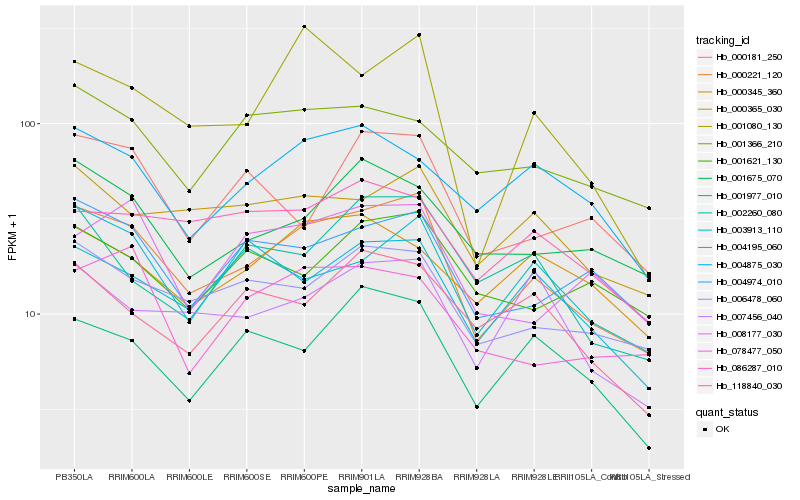
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000221_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 2 |
Hb_006478_060 |
0.1079694688 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004974_010 |
0.116737143 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Prunus mume] |
| 4 |
Hb_000365_030 |
0.1235072662 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 5 |
Hb_001080_130 |
0.1284014991 |
- |
- |
catalase [Hevea brasiliensis] |
| 6 |
Hb_001675_070 |
0.1288957867 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 7 |
Hb_001977_010 |
0.1296974153 |
- |
- |
PREDICTED: uncharacterized protein LOC105636485 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001366_210 |
0.1329712645 |
- |
- |
cathepsin B, putative [Ricinus communis] |
| 9 |
Hb_007456_040 |
0.1332614694 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |
| 10 |
Hb_004875_030 |
0.1349771702 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Jatropha curcas] |
| 11 |
Hb_004195_060 |
0.1349941937 |
- |
- |
PREDICTED: uncharacterized protein LOC105803780 isoform X1 [Gossypium raimondii] |
| 12 |
Hb_008177_030 |
0.135205247 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_086287_010 |
0.135247936 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000181_250 |
0.1371802369 |
- |
- |
PREDICTED: probable bifunctional methylthioribulose-1-phosphate dehydratase/enolase-phosphatase E1 [Jatropha curcas] |
| 15 |
Hb_078477_050 |
0.1380247506 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105648332 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003913_110 |
0.1385824327 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 17 |
Hb_002260_080 |
0.1388453665 |
- |
- |
Aldehyde dehydrogenase family 6 member B2 [Morus notabilis] |
| 18 |
Hb_118840_030 |
0.1400259718 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 18 homolog [Jatropha curcas] |
| 19 |
Hb_001621_130 |
0.1419313077 |
- |
- |
PREDICTED: pumilio homolog 1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000345_360 |
0.1427853224 |
- |
- |
PREDICTED: protein AUXIN RESPONSE 4 [Jatropha curcas] |