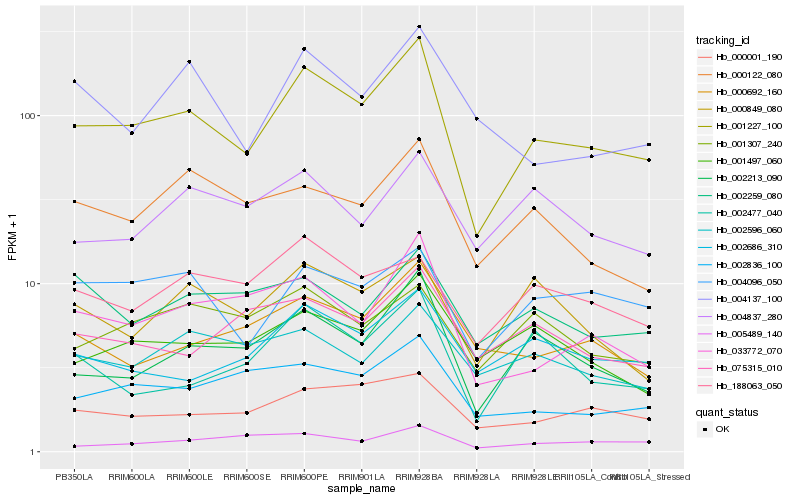
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001227_100 |
0.0 |
- |
- |
RecName: Full=Enolase 1; AltName: Full=2-phospho-D-glycerate hydro-lyase 1; AltName: Full=2-phosphoglycerate dehydratase 1; AltName: Allergen=Hev b 9 [Hevea brasiliensis] |
| 2 |
Hb_002213_090 |
0.1193869031 |
- |
- |
PREDICTED: uncharacterized protein LOC105640147 [Jatropha curcas] |
| 3 |
Hb_033772_070 |
0.1308068551 |
- |
- |
PREDICTED: uncharacterized protein LOC105633681 [Jatropha curcas] |
| 4 |
Hb_000001_190 |
0.1326919606 |
- |
- |
hypothetical protein CISIN_1g0082522mg, partial [Citrus sinensis] |
| 5 |
Hb_002836_100 |
0.1360296144 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004096_050 |
0.1364387799 |
- |
- |
hypothetical protein B456_006G229700 [Gossypium raimondii] |
| 7 |
Hb_001497_060 |
0.1443539034 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 8 |
Hb_002686_310 |
0.1450409594 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 9 |
Hb_000692_160 |
0.1466568256 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 10 |
Hb_000122_080 |
0.1470966253 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |
| 11 |
Hb_002477_040 |
0.1503760871 |
- |
- |
P-loop containing nucleoside triphosphate hydrolases superfamily protein isoform 2 [Theobroma cacao] |
| 12 |
Hb_001307_240 |
0.151355898 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000849_080 |
0.1518784219 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKU5 isoform X1 [Jatropha curcas] |
| 14 |
Hb_188063_050 |
0.1522136403 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 15 |
Hb_004137_100 |
0.1545284876 |
- |
- |
PREDICTED: alcohol dehydrogenase 1 [Jatropha curcas] |
| 16 |
Hb_002596_060 |
0.1545330715 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005489_140 |
0.1554856256 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 18 |
Hb_075315_010 |
0.155551489 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 19 |
Hb_002259_080 |
0.1571744468 |
- |
- |
protein transport protein sec23, putative [Ricinus communis] |
| 20 |
Hb_004837_280 |
0.1586676545 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |