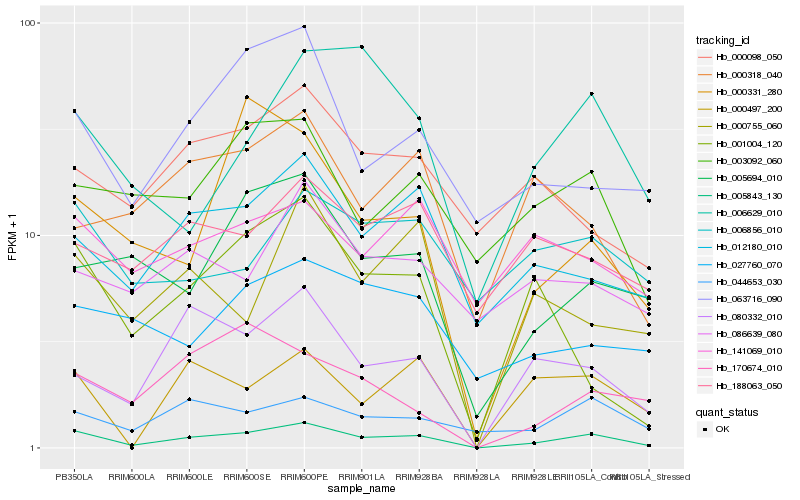
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005843_130 |
0.0 |
- |
- |
hypothetical protein JCGZ_12389 [Jatropha curcas] |
| 2 |
Hb_080332_010 |
0.1994472817 |
- |
- |
hypothetical protein POPTR_0018s10590g [Populus trichocarpa] |
| 3 |
Hb_012180_010 |
0.2048297445 |
- |
- |
PREDICTED: (S)-ureidoglycine aminohydrolase [Jatropha curcas] |
| 4 |
Hb_000497_200 |
0.2103289843 |
- |
- |
- |
| 5 |
Hb_044653_030 |
0.2148862146 |
- |
- |
- |
| 6 |
Hb_063716_090 |
0.2154913525 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000755_060 |
0.2164917052 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104424740 [Eucalyptus grandis] |
| 8 |
Hb_005694_010 |
0.2168039141 |
transcription factor |
TF Family: C2C2-Dof |
F-box family protein, putative isoform 1 [Theobroma cacao] |
| 9 |
Hb_003092_060 |
0.2193252475 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 10 |
Hb_001004_120 |
0.222304037 |
- |
- |
PREDICTED: receptor-like protein kinase HERK 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000331_280 |
0.2295870403 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 12 |
Hb_000098_050 |
0.231850654 |
- |
- |
BnaCnng11900D [Brassica napus] |
| 13 |
Hb_006629_010 |
0.233455918 |
- |
- |
Glutathione S-transferase 2 isoform 3 [Theobroma cacao] |
| 14 |
Hb_188063_050 |
0.2337749401 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 15 |
Hb_006856_010 |
0.2365880754 |
- |
- |
hypothetical protein CISIN_1g019592mg [Citrus sinensis] |
| 16 |
Hb_000318_040 |
0.2368646011 |
- |
- |
hypothetical protein JCGZ_04877 [Jatropha curcas] |
| 17 |
Hb_086639_080 |
0.2371842546 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_170674_010 |
0.2376823318 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |
| 19 |
Hb_027760_070 |
0.2383484154 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 20 |
Hb_141069_010 |
0.2385276847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |