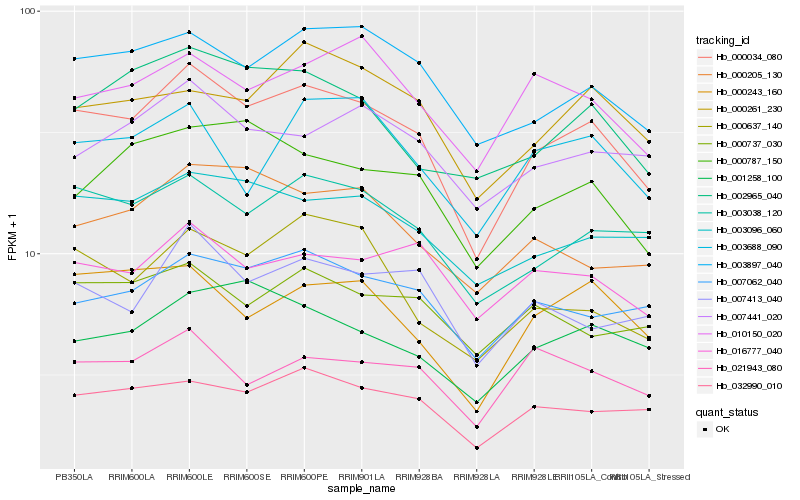
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000034_080 |
0.0 |
- |
- |
PREDICTED: MADS-box protein AGL24-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_003038_120 |
0.0806543425 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 3 |
Hb_003897_040 |
0.0825948234 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 4 |
Hb_032990_010 |
0.0868226238 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 5 |
Hb_003096_060 |
0.0887017501 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 6 |
Hb_007062_040 |
0.0914845095 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 7 |
Hb_007413_040 |
0.0952183534 |
- |
- |
myo inositol monophosphatase, putative [Ricinus communis] |
| 8 |
Hb_000787_150 |
0.095255492 |
- |
- |
PREDICTED: protein NDR1-like [Jatropha curcas] |
| 9 |
Hb_000243_160 |
0.0961982578 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 10 |
Hb_002965_040 |
0.0971686232 |
- |
- |
drought-responsive family protein [Populus trichocarpa] |
| 11 |
Hb_000261_230 |
0.0983700179 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |
| 12 |
Hb_016777_040 |
0.0990917205 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 13 |
Hb_000637_140 |
0.0998837452 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 14 |
Hb_001258_100 |
0.1000275322 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_010150_020 |
0.1003117941 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 16 |
Hb_007441_020 |
0.1006699763 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 17 |
Hb_000205_130 |
0.1012997826 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 18 |
Hb_000737_030 |
0.101831036 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003688_090 |
0.1037269817 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 20 |
Hb_021943_080 |
0.1041708057 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |