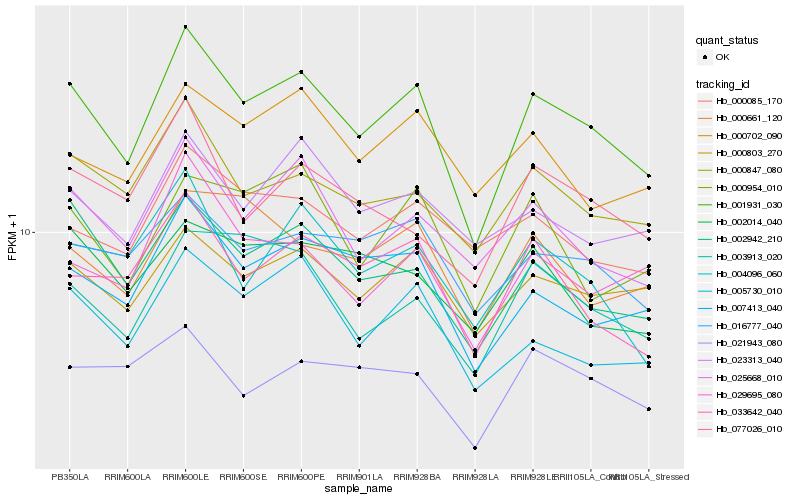
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001931_030 |
0.0 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta-like [Nelumbo nucifera] |
| 2 |
Hb_007413_040 |
0.0771839929 |
- |
- |
myo inositol monophosphatase, putative [Ricinus communis] |
| 3 |
Hb_002942_210 |
0.0779086869 |
- |
- |
alpha-2,8-sialyltransferase 8b, putative [Ricinus communis] |
| 4 |
Hb_005730_010 |
0.0810830976 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 5 |
Hb_025668_010 |
0.0811318521 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_000803_270 |
0.0812998585 |
- |
- |
PREDICTED: nuclear cap-binding protein subunit 1 [Jatropha curcas] |
| 7 |
Hb_000847_080 |
0.0828140519 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 8 |
Hb_033642_040 |
0.0843951985 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_016777_040 |
0.0868696175 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 10 |
Hb_003913_020 |
0.0881507877 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At5g50170 [Jatropha curcas] |
| 11 |
Hb_029695_080 |
0.0899739958 |
- |
- |
PREDICTED: NAD-dependent protein deacetylase SRT1 [Prunus mume] |
| 12 |
Hb_000954_010 |
0.092471247 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 13 |
Hb_004096_060 |
0.0930688065 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase isoform X2 [Jatropha curcas] |
| 14 |
Hb_023313_040 |
0.0943694331 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000661_120 |
0.09469133 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 16 |
Hb_000702_090 |
0.0961820336 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 17 |
Hb_021943_080 |
0.0963453722 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_077026_010 |
0.0968912615 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 2 [Jatropha curcas] |
| 19 |
Hb_000085_170 |
0.0979369733 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_002014_040 |
0.0997893049 |
- |
- |
site-1 protease, putative [Ricinus communis] |