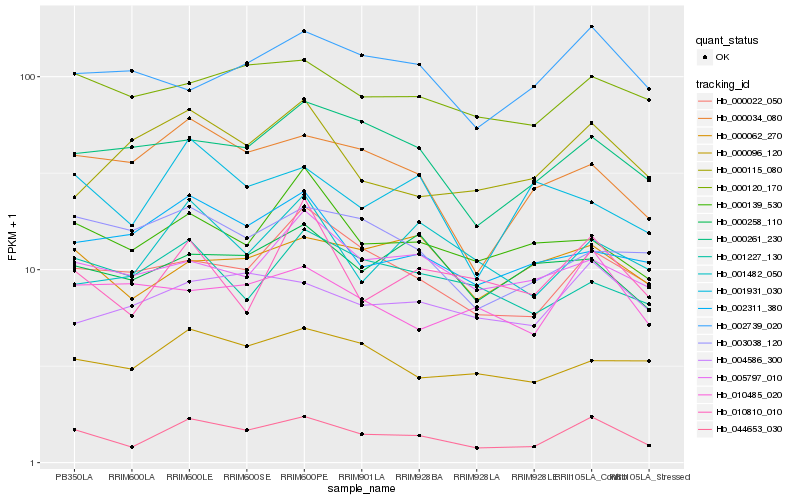
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_044653_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_000022_050 |
0.1274128531 |
- |
- |
PREDICTED: retinol dehydrogenase 11-like isoform X2 [Glycine max] |
| 3 |
Hb_000261_230 |
0.1346501547 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |
| 4 |
Hb_000034_080 |
0.1349959646 |
- |
- |
PREDICTED: MADS-box protein AGL24-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000120_170 |
0.1360052562 |
- |
- |
selenoprotein [Populus trichocarpa] |
| 6 |
Hb_004586_300 |
0.1367941158 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000096_120 |
0.1396507418 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 8 |
Hb_010810_010 |
0.1400367606 |
- |
- |
PREDICTED: plastidic glucose transporter 4 [Jatropha curcas] |
| 9 |
Hb_000062_270 |
0.1413487477 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_010485_020 |
0.1423360023 |
- |
- |
PREDICTED: thaumatin-like protein 1b isoform X1 [Jatropha curcas] |
| 11 |
Hb_000139_530 |
0.1433629411 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005797_010 |
0.1443229067 |
- |
- |
PREDICTED: lysophospholipid acyltransferase 1-like [Jatropha curcas] |
| 13 |
Hb_002739_020 |
0.1471579168 |
- |
- |
PREDICTED: UTP--glucose-1-phosphate uridylyltransferase [Jatropha curcas] |
| 14 |
Hb_002311_380 |
0.147171666 |
- |
- |
PREDICTED: induced during hyphae development protein 1 [Jatropha curcas] |
| 15 |
Hb_000258_110 |
0.1474412021 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001227_130 |
0.1495049784 |
- |
- |
PREDICTED: delta(14)-sterol reductase [Jatropha curcas] |
| 17 |
Hb_001482_050 |
0.1496552366 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 18 |
Hb_003038_120 |
0.1522444392 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 19 |
Hb_001931_030 |
0.1524994361 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta-like [Nelumbo nucifera] |
| 20 |
Hb_000115_080 |
0.1526228091 |
- |
- |
PREDICTED: uncharacterized protein LOC105642032 [Jatropha curcas] |