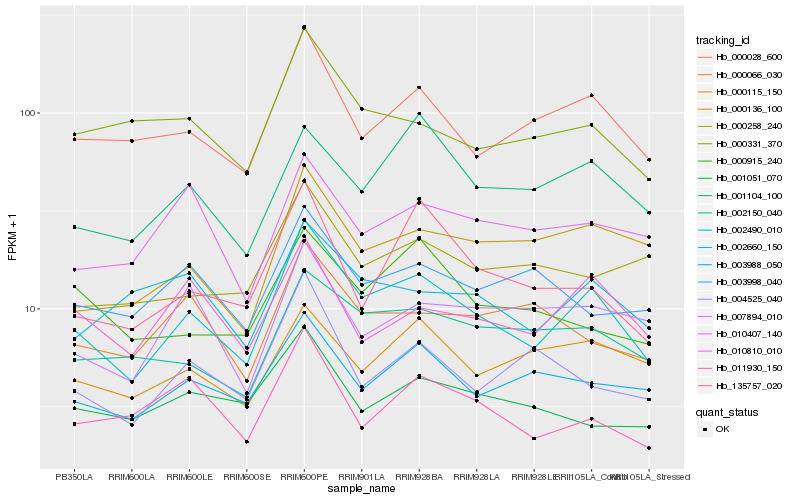
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002490_010 |
0.0 |
- |
- |
PREDICTED: plastidic glucose transporter 4 [Jatropha curcas] |
| 2 |
Hb_000028_600 |
0.112004263 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 3 |
Hb_003988_050 |
0.1265687469 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 4 |
Hb_002150_040 |
0.1276668176 |
- |
- |
PREDICTED: CMP-sialic acid transporter 1 isoform X3 [Jatropha curcas] |
| 5 |
Hb_010810_010 |
0.1334834213 |
- |
- |
PREDICTED: plastidic glucose transporter 4 [Jatropha curcas] |
| 6 |
Hb_000136_100 |
0.1336760677 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 7 |
Hb_001104_100 |
0.1339093658 |
- |
- |
RecName: Full=Enolase 2; AltName: Full=2-phospho-D-glycerate hydro-lyase 2; AltName: Full=2-phosphoglycerate dehydratase 2; AltName: Allergen=Hev b 9 [Hevea brasiliensis] |
| 8 |
Hb_000331_370 |
0.1352098464 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 9 |
Hb_002660_150 |
0.1376269736 |
- |
- |
PREDICTED: uncharacterized protein LOC105642207 isoform X1 [Jatropha curcas] |
| 10 |
Hb_011930_150 |
0.138567524 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 11 |
Hb_135757_020 |
0.1386528506 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 12 |
Hb_000115_150 |
0.1393374623 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 13 |
Hb_004525_040 |
0.1432167751 |
- |
- |
PREDICTED: sugar transporter ERD6-like 6 [Jatropha curcas] |
| 14 |
Hb_010407_140 |
0.1432796304 |
- |
- |
PREDICTED: malate dehydrogenase [Jatropha curcas] |
| 15 |
Hb_007894_010 |
0.1437582845 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 16 |
Hb_000066_030 |
0.1438048021 |
- |
- |
PREDICTED: uncharacterized protein LOC105643898 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000915_240 |
0.144523432 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_001051_070 |
0.1457491702 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000258_240 |
0.1461994169 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 20 |
Hb_003998_040 |
0.1465061226 |
- |
- |
organic anion transporter, putative [Ricinus communis] |