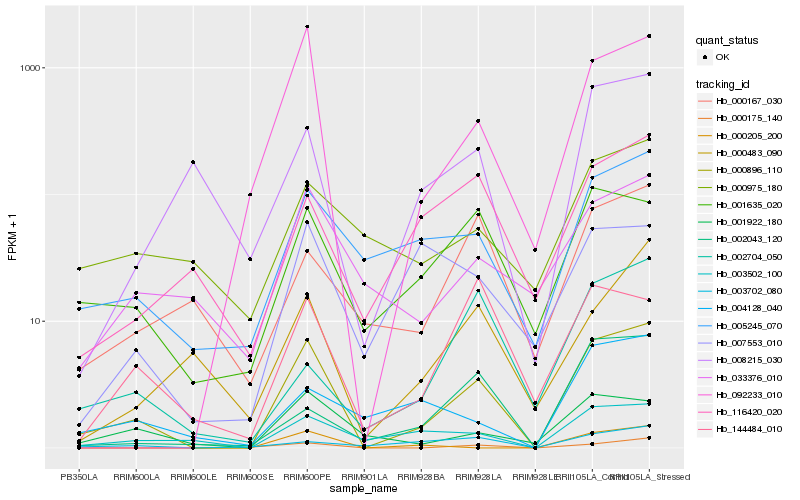
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000896_110 |
0.0 |
- |
- |
- |
| 2 |
Hb_092233_010 |
0.2126280156 |
- |
- |
RecName: Full=Profilin-3; AltName: Full=Pollen allergen Hev b 8.0201; AltName: Allergen=Hev b 8.0201 [Hevea brasiliensis] |
| 3 |
Hb_005245_070 |
0.2341987845 |
- |
- |
PREDICTED: uncharacterized protein LOC105643658 [Jatropha curcas] |
| 4 |
Hb_000175_140 |
0.2358934744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001635_020 |
0.2420220841 |
- |
- |
PREDICTED: inorganic pyrophosphatase 1-like [Jatropha curcas] |
| 6 |
Hb_003502_100 |
0.2436379206 |
- |
- |
- |
| 7 |
Hb_001922_180 |
0.24505241 |
- |
- |
hevamine-A precursor, putative [Ricinus communis] |
| 8 |
Hb_033376_010 |
0.252220074 |
- |
- |
hypothetical protein POPTR_0008s02960g [Populus trichocarpa] |
| 9 |
Hb_008215_030 |
0.2585306386 |
- |
- |
hypothetical protein 3 [Hevea brasiliensis] |
| 10 |
Hb_000483_090 |
0.267124279 |
- |
- |
- |
| 11 |
Hb_003702_080 |
0.2695465284 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 12 |
Hb_002043_120 |
0.2718658381 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_116420_020 |
0.2738282365 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 14 |
Hb_000205_200 |
0.2750726767 |
- |
- |
- |
| 15 |
Hb_144484_010 |
0.2754026757 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 16 |
Hb_004128_040 |
0.2778088529 |
- |
- |
- |
| 17 |
Hb_000167_030 |
0.2779783628 |
- |
- |
PREDICTED: uncharacterized protein LOC105641111 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000975_180 |
0.2815814751 |
- |
- |
Uncharacterized protein TCM_041879 [Theobroma cacao] |
| 19 |
Hb_007553_010 |
0.2892044459 |
- |
- |
- |
| 20 |
Hb_002704_050 |
0.2897567644 |
- |
- |
cysteine-type peptidase, putative [Ricinus communis] |