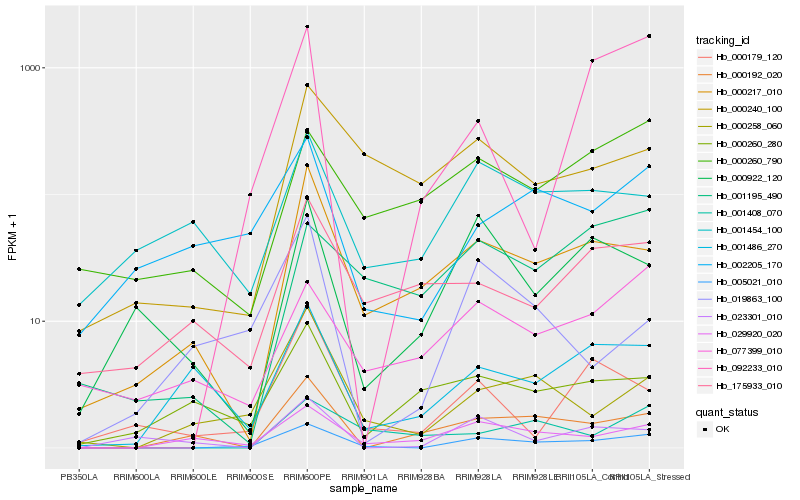
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005021_010 |
0.0 |
- |
- |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 2 |
Hb_029920_020 |
0.2459019318 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG7 [Jatropha curcas] |
| 3 |
Hb_000258_060 |
0.2467072739 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000217_010 |
0.2568560392 |
- |
- |
hypothetical protein POPTR_0341s00210g [Populus trichocarpa] |
| 5 |
Hb_002205_170 |
0.2688903879 |
- |
- |
PREDICTED: uncharacterized protein LOC105649179 [Jatropha curcas] |
| 6 |
Hb_001486_270 |
0.27577499 |
- |
- |
Ann10 [Manihot esculenta] |
| 7 |
Hb_092233_010 |
0.2785096728 |
- |
- |
RecName: Full=Profilin-3; AltName: Full=Pollen allergen Hev b 8.0201; AltName: Allergen=Hev b 8.0201 [Hevea brasiliensis] |
| 8 |
Hb_000192_020 |
0.2793651152 |
- |
- |
hypothetical protein JCGZ_17351 [Jatropha curcas] |
| 9 |
Hb_000240_100 |
0.282280083 |
- |
- |
PREDICTED: ras-related protein Rab5-like [Eucalyptus grandis] |
| 10 |
Hb_077399_010 |
0.2922569228 |
- |
- |
hypothetical protein POPTR_0003s09910g [Populus trichocarpa] |
| 11 |
Hb_001408_070 |
0.2929068147 |
- |
- |
- |
| 12 |
Hb_000922_120 |
0.2942479654 |
- |
- |
PREDICTED: carbonic anhydrase 2-like [Jatropha curcas] |
| 13 |
Hb_000260_790 |
0.2952643438 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 12 isoform 1 [Theobroma cacao] |
| 14 |
Hb_000179_120 |
0.2970884591 |
- |
- |
PREDICTED: corepressor interacting with RBPJ 1-like [Jatropha curcas] |
| 15 |
Hb_175933_010 |
0.3015399478 |
- |
- |
NADH-ubiquinone oxidoreductase 24 kD subunit, putative [Ricinus communis] |
| 16 |
Hb_000260_280 |
0.3031196869 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 17 |
Hb_001195_490 |
0.303125161 |
- |
- |
hypothetical protein glysoja_018593 [Glycine soja] |
| 18 |
Hb_001454_100 |
0.3037769655 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 19 |
Hb_023301_010 |
0.3046175208 |
- |
- |
PREDICTED: serine/threonine-protein kinase AGC1-7 [Jatropha curcas] |
| 20 |
Hb_019863_100 |
0.3075326184 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 2, mitochondrial-like [Jatropha curcas] |