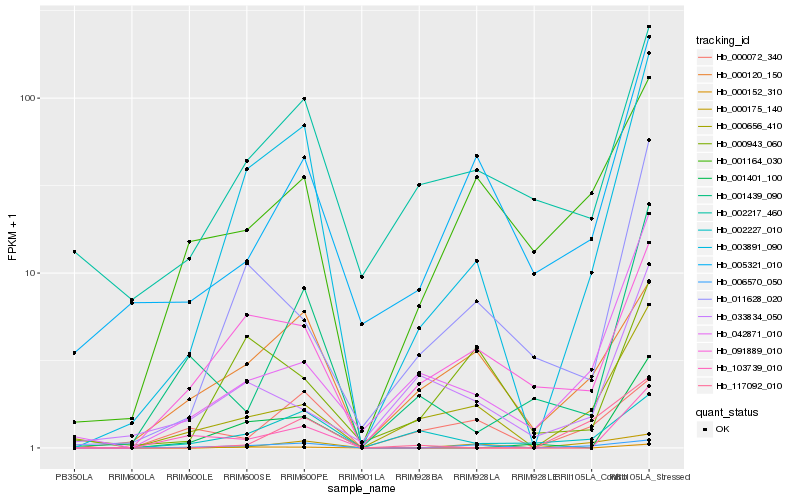
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003891_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_000152_310 |
0.2325030726 |
- |
- |
- |
| 3 |
Hb_001401_100 |
0.238141205 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 4 |
Hb_091889_010 |
0.2452694107 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_002227_010 |
0.2509797219 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Populus euphratica] |
| 6 |
Hb_000656_410 |
0.258282699 |
- |
- |
hypothetical protein POPTR_0003s06410g [Populus trichocarpa] |
| 7 |
Hb_011628_020 |
0.2585807461 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006570_050 |
0.2596971635 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 9 |
Hb_000120_150 |
0.2636155404 |
- |
- |
hypothetical protein JCGZ_08453 [Jatropha curcas] |
| 10 |
Hb_002217_460 |
0.2684436617 |
- |
- |
PREDICTED: transcription factor bHLH147-like [Jatropha curcas] |
| 11 |
Hb_042871_010 |
0.2714175471 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 12 |
Hb_001439_090 |
0.2790218753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000943_060 |
0.282424831 |
- |
- |
PREDICTED: uncharacterized protein LOC105635589 [Jatropha curcas] |
| 14 |
Hb_005321_010 |
0.2832783294 |
- |
- |
PREDICTED: uncharacterized protein LOC105644542 [Jatropha curcas] |
| 15 |
Hb_103739_010 |
0.2842072272 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 16 |
Hb_001164_030 |
0.3035473924 |
- |
- |
GMP synthase [glutamine-hydrolyzing] subunit A, putative [Ricinus communis] |
| 17 |
Hb_000175_140 |
0.3071568866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_117092_010 |
0.3106323954 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 19 |
Hb_033834_050 |
0.3147343033 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000072_340 |
0.3174862223 |
- |
- |
conserved hypothetical protein [Ricinus communis] |