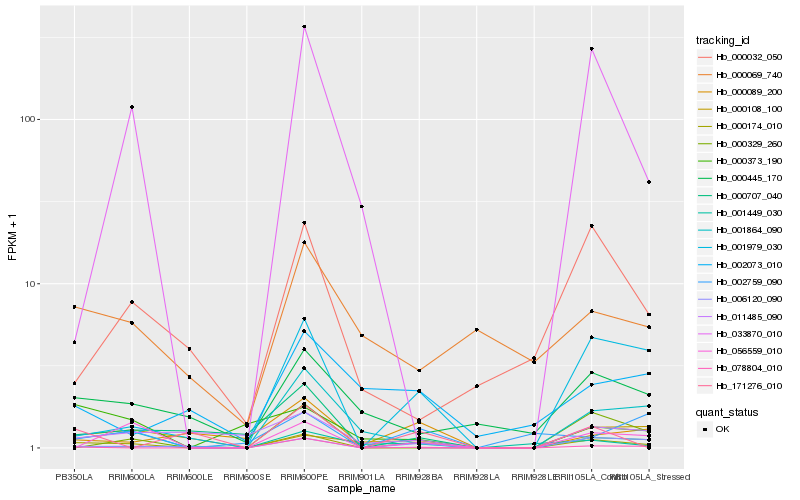
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000174_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103444904 [Malus domestica] |
| 2 |
Hb_006120_090 |
0.2780172274 |
- |
- |
PREDICTED: desiccation-related protein PCC13-62-like [Jatropha curcas] |
| 3 |
Hb_001979_030 |
0.3012955228 |
- |
- |
hypothetical protein POPTR_0016s06130g [Populus trichocarpa] |
| 4 |
Hb_078804_010 |
0.3415935551 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_001864_090 |
0.3465608023 |
- |
- |
- |
| 6 |
Hb_056559_010 |
0.3485755058 |
- |
- |
PREDICTED: uncharacterized protein LOC104088595 [Nicotiana tomentosiformis] |
| 7 |
Hb_000445_170 |
0.3619786387 |
- |
- |
PREDICTED: probable methyltransferase PMT10 [Jatropha curcas] |
| 8 |
Hb_000089_200 |
0.3664105663 |
- |
- |
hypothetical protein VITISV_019292 [Vitis vinifera] |
| 9 |
Hb_002759_090 |
0.3719699639 |
- |
- |
- |
| 10 |
Hb_000373_190 |
0.3726574517 |
- |
- |
- |
| 11 |
Hb_011485_090 |
0.3729504007 |
- |
- |
- |
| 12 |
Hb_000707_040 |
0.3848091343 |
- |
- |
Uncharacterized protein TCM_021829 [Theobroma cacao] |
| 13 |
Hb_000069_740 |
0.386678175 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 14 |
Hb_000108_100 |
0.3886775266 |
- |
- |
PREDICTED: DNA polymerase epsilon subunit 2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000032_050 |
0.3897543804 |
- |
- |
hypothetical protein RCOM_1691130 [Ricinus communis] |
| 16 |
Hb_000329_260 |
0.3900133677 |
- |
- |
hypothetical protein MIMGU_mgv1a016162mg [Erythranthe guttata] |
| 17 |
Hb_033870_010 |
0.3921003808 |
- |
- |
PREDICTED: uncharacterized protein LOC104101109 isoform X1 [Nicotiana tomentosiformis] |
| 18 |
Hb_171276_010 |
0.3938255488 |
- |
- |
- |
| 19 |
Hb_001449_030 |
0.3955286718 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X3 [Jatropha curcas] |
| 20 |
Hb_002073_010 |
0.3975236544 |
- |
- |
- |