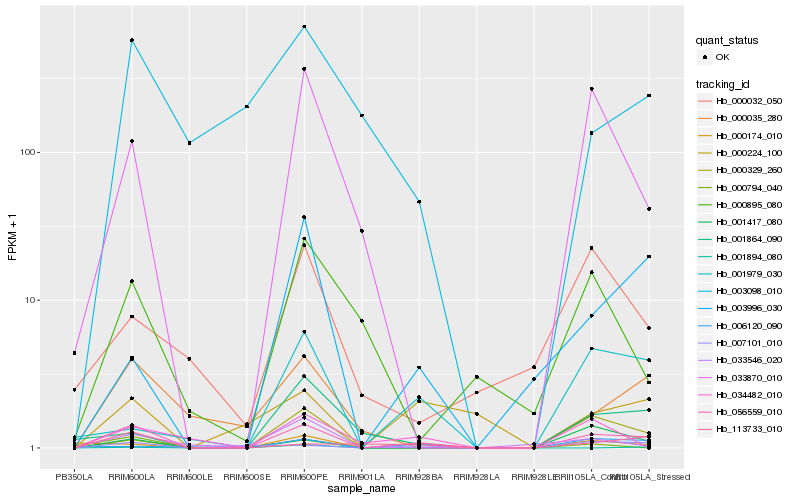
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_056559_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104088595 [Nicotiana tomentosiformis] |
| 2 |
Hb_007101_010 |
0.3247403663 |
- |
- |
PREDICTED: 2-hydroxyisoflavanone dehydratase-like [Jatropha curcas] |
| 3 |
Hb_033870_010 |
0.3333147414 |
- |
- |
PREDICTED: uncharacterized protein LOC104101109 isoform X1 [Nicotiana tomentosiformis] |
| 4 |
Hb_000329_260 |
0.3410509501 |
- |
- |
hypothetical protein MIMGU_mgv1a016162mg [Erythranthe guttata] |
| 5 |
Hb_113733_010 |
0.3438221225 |
- |
- |
hypothetical protein JCGZ_26531 [Jatropha curcas] |
| 6 |
Hb_000035_280 |
0.3448236914 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230-like [Jatropha curcas] |
| 7 |
Hb_000174_010 |
0.3485755058 |
- |
- |
PREDICTED: uncharacterized protein LOC103444904 [Malus domestica] |
| 8 |
Hb_001417_080 |
0.350052342 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 9 |
Hb_000224_100 |
0.3637316916 |
- |
- |
- |
| 10 |
Hb_001979_030 |
0.3655475833 |
- |
- |
hypothetical protein POPTR_0016s06130g [Populus trichocarpa] |
| 11 |
Hb_033546_020 |
0.3691810873 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor pho1, putative [Ricinus communis] |
| 12 |
Hb_003996_030 |
0.3703948401 |
- |
- |
- |
| 13 |
Hb_000794_040 |
0.3706677723 |
- |
- |
PREDICTED: RING-H2 finger protein ATL29-like [Jatropha curcas] |
| 14 |
Hb_001864_090 |
0.3741856776 |
- |
- |
- |
| 15 |
Hb_003098_010 |
0.3765321193 |
- |
- |
- |
| 16 |
Hb_034482_010 |
0.3816913673 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000895_080 |
0.3819485615 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1-like [Jatropha curcas] |
| 18 |
Hb_006120_090 |
0.3824699097 |
- |
- |
PREDICTED: desiccation-related protein PCC13-62-like [Jatropha curcas] |
| 19 |
Hb_001894_080 |
0.3889023429 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_000032_050 |
0.3926105894 |
- |
- |
hypothetical protein RCOM_1691130 [Ricinus communis] |