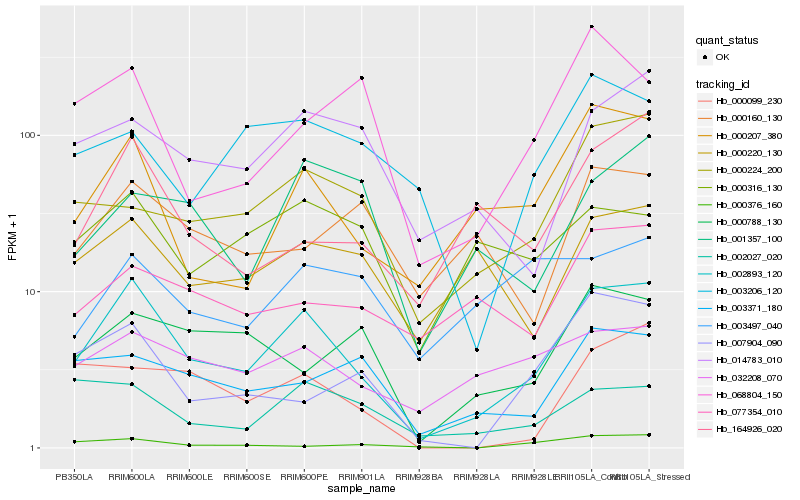
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002893_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637692 [Jatropha curcas] |
| 2 |
Hb_000207_380 |
0.1905793822 |
- |
- |
sucrose transporter 6 [Hevea brasiliensis] |
| 3 |
Hb_032208_070 |
0.2147614484 |
- |
- |
PREDICTED: probable polyamine transporter At3g13620 isoform X2 [Jatropha curcas] |
| 4 |
Hb_014783_010 |
0.2156713792 |
- |
- |
cold-inducible RNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_000224_200 |
0.2185204406 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 6 |
Hb_164926_020 |
0.2230629898 |
- |
- |
PREDICTED: uncharacterized protein LOC105645601 [Jatropha curcas] |
| 7 |
Hb_003371_180 |
0.2280552378 |
- |
- |
- |
| 8 |
Hb_000220_130 |
0.2296299435 |
- |
- |
Phosphatidyl serine synthase family protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_000099_230 |
0.2297853829 |
- |
- |
PREDICTED: F-box protein SKIP19-like [Gossypium raimondii] |
| 10 |
Hb_007904_090 |
0.2310861737 |
- |
- |
PREDICTED: uncharacterized protein LOC100261196 [Vitis vinifera] |
| 11 |
Hb_002027_020 |
0.2312794335 |
- |
- |
PREDICTED: uncharacterized protein LOC105650039 [Jatropha curcas] |
| 12 |
Hb_077354_010 |
0.233645988 |
- |
- |
PREDICTED: uncharacterized protein LOC105637567 [Jatropha curcas] |
| 13 |
Hb_000788_130 |
0.235779081 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 14 |
Hb_001357_100 |
0.2388313868 |
- |
- |
PREDICTED: small ubiquitin-related modifier 1 [Jatropha curcas] |
| 15 |
Hb_000316_130 |
0.2420572855 |
- |
- |
PREDICTED: uncharacterized protein LOC105647693 [Jatropha curcas] |
| 16 |
Hb_000376_160 |
0.2447590318 |
- |
- |
- |
| 17 |
Hb_003206_120 |
0.245842048 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |
| 18 |
Hb_068804_150 |
0.2472998343 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000160_130 |
0.2511760289 |
- |
- |
PREDICTED: uncharacterized protein LOC105648215 [Jatropha curcas] |
| 20 |
Hb_003497_040 |
0.251212469 |
- |
- |
ATP binding protein, putative [Ricinus communis] |