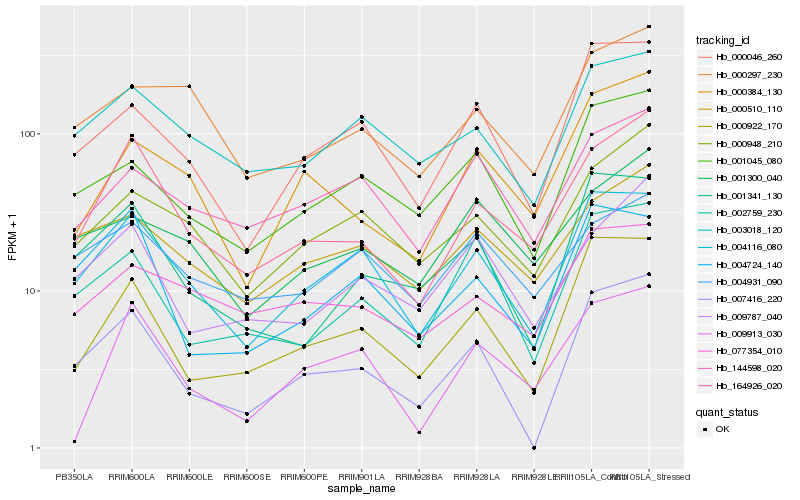
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_164926_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645601 [Jatropha curcas] |
| 2 |
Hb_004116_080 |
0.143110807 |
- |
- |
ATATH9, putative [Ricinus communis] |
| 3 |
Hb_000384_130 |
0.1431911098 |
- |
- |
uridylate kinase plant, putative [Ricinus communis] |
| 4 |
Hb_009787_040 |
0.1444961514 |
desease resistance |
Gene Name: AAA_17 |
PREDICTED: uncharacterized protein LOC105135462 isoform X2 [Populus euphratica] |
| 5 |
Hb_000948_210 |
0.1455292941 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000922_170 |
0.1536675378 |
- |
- |
PREDICTED: uncharacterized protein LOC105640383 [Jatropha curcas] |
| 7 |
Hb_000510_110 |
0.1591978217 |
- |
- |
PREDICTED: signal recognition particle 9 kDa protein [Jatropha curcas] |
| 8 |
Hb_003018_120 |
0.1601942801 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein F [Jatropha curcas] |
| 9 |
Hb_000297_230 |
0.1608647761 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 10 |
Hb_009913_030 |
0.1649621923 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein JCGZ_05054 [Jatropha curcas] |
| 11 |
Hb_077354_010 |
0.1660486224 |
- |
- |
PREDICTED: uncharacterized protein LOC105637567 [Jatropha curcas] |
| 12 |
Hb_007416_220 |
0.1667612949 |
- |
- |
PREDICTED: uncharacterized protein LOC105637482 [Jatropha curcas] |
| 13 |
Hb_001341_130 |
0.1684469162 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001300_040 |
0.1708161863 |
- |
- |
PREDICTED: uncharacterized protein LOC105642466 [Jatropha curcas] |
| 15 |
Hb_004724_140 |
0.1711282684 |
transcription factor |
TF Family: NF-YB |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 16 |
Hb_144598_020 |
0.1735084002 |
- |
- |
PREDICTED: triphosphate tunel metalloenzyme 3 [Jatropha curcas] |
| 17 |
Hb_000046_260 |
0.1760496351 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 18 |
Hb_002759_230 |
0.1770015641 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 25 [Jatropha curcas] |
| 19 |
Hb_001045_080 |
0.1770989508 |
- |
- |
PREDICTED: uncharacterized protein LOC105646704 [Jatropha curcas] |
| 20 |
Hb_004931_090 |
0.1787191363 |
- |
- |
PREDICTED: chromatin modification-related protein MEAF6-like isoform X2 [Jatropha curcas] |