| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
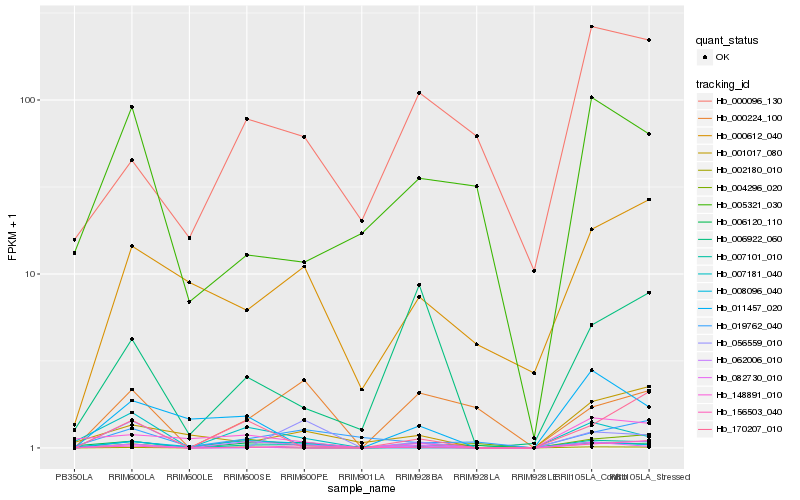
Hb_007101_010 |
0.0 |
- |
- |
PREDICTED: 2-hydroxyisoflavanone dehydratase-like [Jatropha curcas] |
| 2 |
Hb_062006_010 |
0.2003853639 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 3 |
Hb_007181_040 |
0.262303186 |
- |
- |
- |
| 4 |
Hb_082730_010 |
0.3044306402 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 5 |
Hb_000612_040 |
0.3109989526 |
- |
- |
hypothetical protein OsJ_16945 [Oryza sativa Japonica Group] |
| 6 |
Hb_000224_100 |
0.3218416512 |
- |
- |
- |
| 7 |
Hb_056559_010 |
0.3247403663 |
- |
- |
PREDICTED: uncharacterized protein LOC104088595 [Nicotiana tomentosiformis] |
| 8 |
Hb_156503_040 |
0.3268476577 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 9 |
Hb_006120_110 |
0.3284800989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001017_080 |
0.3290141151 |
- |
- |
hypothetical protein POPTR_0010s06020g [Populus trichocarpa] |
| 11 |
Hb_006922_060 |
0.3296157689 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 12 |
Hb_148891_010 |
0.3296426107 |
- |
- |
- |
| 13 |
Hb_019762_040 |
0.332457973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002180_010 |
0.3391196457 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 15 |
Hb_170207_010 |
0.342172413 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 16 |
Hb_011457_020 |
0.3457669656 |
- |
- |
PREDICTED: uncharacterized protein LOC105649960 [Jatropha curcas] |
| 17 |
Hb_000096_130 |
0.3472970401 |
- |
- |
hypothetical protein 23 [Hevea brasiliensis] |
| 18 |
Hb_005321_030 |
0.3480453718 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4 [Jatropha curcas] |
| 19 |
Hb_008096_040 |
0.3511229479 |
- |
- |
hypothetical protein PRUPE_ppa001351mg [Prunus persica] |
| 20 |
Hb_004296_020 |
0.3517656909 |
- |
- |
unnamed protein product [Coffea canephora] |