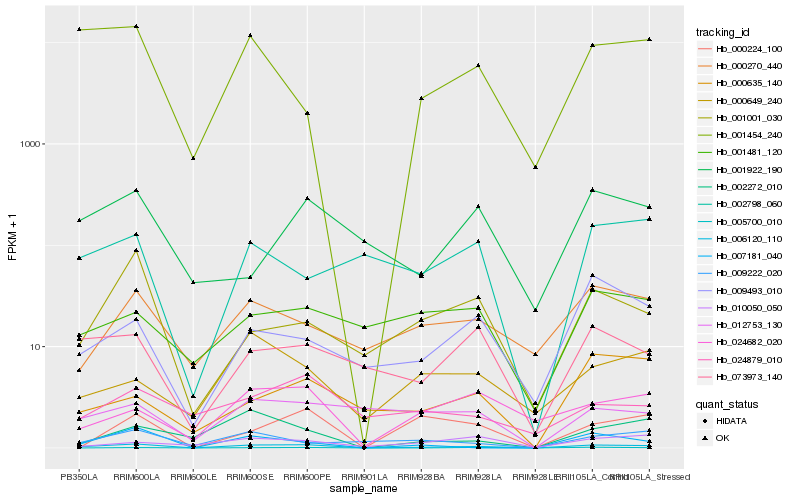
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006120_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007181_040 |
0.2670764402 |
- |
- |
- |
| 3 |
Hb_073973_140 |
0.2725519446 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 4 |
Hb_001454_240 |
0.2742594214 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 5 |
Hb_024682_020 |
0.2765391459 |
- |
- |
hypothetical protein POPTR_0001s12710g [Populus trichocarpa] |
| 6 |
Hb_002272_010 |
0.2786730761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000270_440 |
0.2873166775 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0015s08130g [Populus trichocarpa] |
| 8 |
Hb_001001_030 |
0.2913074551 |
- |
- |
hypothetical protein POPTR_0014s03230g [Populus trichocarpa] |
| 9 |
Hb_009493_010 |
0.2924823082 |
- |
- |
Minor allergen Alt a, putative [Ricinus communis] |
| 10 |
Hb_012753_130 |
0.2953023407 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000649_240 |
0.297850619 |
- |
- |
PREDICTED: probable peroxygenase 4 [Jatropha curcas] |
| 12 |
Hb_009222_020 |
0.2979279926 |
- |
- |
hypothetical protein AALP_AA5G087000 [Arabis alpina] |
| 13 |
Hb_000224_100 |
0.2988348041 |
- |
- |
- |
| 14 |
Hb_002798_060 |
0.3001336519 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 1-like [Vitis vinifera] |
| 15 |
Hb_005700_010 |
0.3008823999 |
- |
- |
PREDICTED: uncharacterized protein LOC104898079 [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_001922_190 |
0.3028094457 |
- |
- |
RecName: Full=Hevamine-A; Includes: RecName: Full=Chitinase; Includes: RecName: Full=Lysozyme; Flags: Precursor [Hevea brasiliensis] |
| 17 |
Hb_024879_010 |
0.3030710388 |
- |
- |
unknown [Glycine max] |
| 18 |
Hb_000635_140 |
0.3045656382 |
transcription factor |
TF Family: NF-YC |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_001481_120 |
0.305569264 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 20 |
Hb_010050_050 |
0.3074833691 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog isoform X2 [Jatropha curcas] |