| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
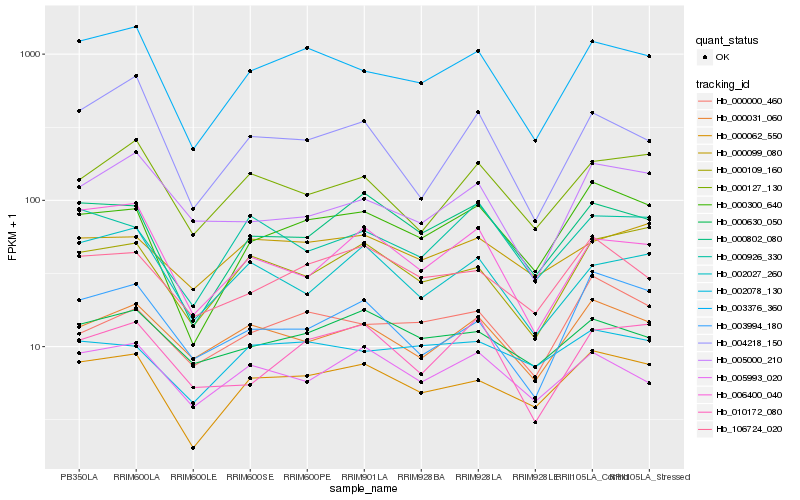
Hb_003376_360 |
0.0 |
- |
- |
PREDICTED: ubiquitin-like protein 5 [Nelumbo nucifera] |
| 2 |
Hb_000062_550 |
0.0922583738 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000300_640 |
0.1098218222 |
- |
- |
PREDICTED: uncharacterized protein LOC100855285 [Vitis vinifera] |
| 4 |
Hb_004218_150 |
0.1139429329 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 5 |
Hb_000031_060 |
0.1150029306 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF12-A-like [Jatropha curcas] |
| 6 |
Hb_000127_130 |
0.1186106039 |
- |
- |
PREDICTED: uncharacterized protein LOC105635483 [Jatropha curcas] |
| 7 |
Hb_006400_040 |
0.1209942762 |
- |
- |
PREDICTED: uncharacterized protein LOC105642527 [Jatropha curcas] |
| 8 |
Hb_000802_080 |
0.1214327257 |
- |
- |
PREDICTED: F-box protein At5g46170-like [Jatropha curcas] |
| 9 |
Hb_106724_020 |
0.123275851 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 1A-like isoform X1 [Populus euphratica] |
| 10 |
Hb_002027_260 |
0.1244905012 |
- |
- |
PREDICTED: gibberellin receptor GID1B [Jatropha curcas] |
| 11 |
Hb_005000_210 |
0.1248998546 |
- |
- |
PREDICTED: translation machinery-associated protein 22 [Jatropha curcas] |
| 12 |
Hb_005993_020 |
0.1273430293 |
- |
- |
PREDICTED: uncharacterized protein LOC105649970 [Jatropha curcas] |
| 13 |
Hb_010172_080 |
0.1284255724 |
- |
- |
acetylglucosaminyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000630_050 |
0.1284774603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000109_160 |
0.1290772619 |
- |
- |
hypothetical protein PRUPE_ppa012786mg [Prunus persica] |
| 16 |
Hb_002078_130 |
0.1291629355 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 17 |
Hb_000000_460 |
0.1294833976 |
- |
- |
PREDICTED: peroxisomal membrane protein 11A [Jatropha curcas] |
| 18 |
Hb_000926_330 |
0.1299555223 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 19 |
Hb_003994_180 |
0.1300469732 |
- |
- |
PREDICTED: probable mitochondrial adenine nucleotide transporter BTL1 [Jatropha curcas] |
| 20 |
Hb_000099_080 |
0.1309602515 |
- |
- |
kinesin light chain, putative [Ricinus communis] |