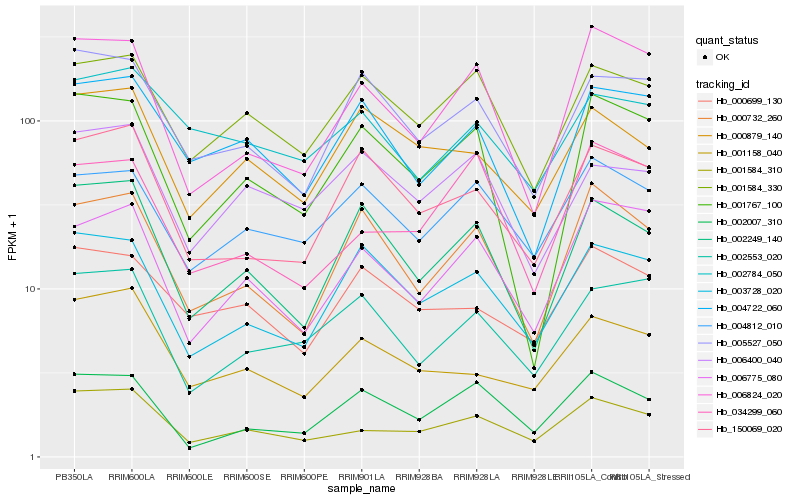
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001584_310 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s00630g [Populus trichocarpa] |
| 2 |
Hb_001158_040 |
0.1062823995 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |
| 3 |
Hb_006824_020 |
0.1206333118 |
- |
- |
PREDICTED: thioredoxin domain-containing protein 9 homolog [Jatropha curcas] |
| 4 |
Hb_006775_080 |
0.1224252525 |
- |
- |
PREDICTED: uncharacterized protein LOC105649730 [Jatropha curcas] |
| 5 |
Hb_001584_330 |
0.1269925792 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha [Jatropha curcas] |
| 6 |
Hb_034299_060 |
0.1281453836 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator ARR5 [Jatropha curcas] |
| 7 |
Hb_002249_140 |
0.1294294682 |
- |
- |
PREDICTED: uncharacterized protein LOC105648715 [Jatropha curcas] |
| 8 |
Hb_004812_010 |
0.1323774652 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD6 [Jatropha curcas] |
| 9 |
Hb_005527_050 |
0.1325947413 |
- |
- |
PREDICTED: uncharacterized protein LOC105643024 [Jatropha curcas] |
| 10 |
Hb_002007_310 |
0.133484912 |
- |
- |
cyclin family protein [Populus trichocarpa] |
| 11 |
Hb_003728_020 |
0.134544729 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002553_020 |
0.1356548357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002784_050 |
0.1373825234 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6 [Jatropha curcas] |
| 14 |
Hb_150069_020 |
0.1380126027 |
- |
- |
PREDICTED: ribosome biogenesis regulatory protein homolog isoform X1 [Jatropha curcas] |
| 15 |
Hb_000879_140 |
0.1382319955 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0283697-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_000699_130 |
0.1394885258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001767_100 |
0.139787362 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 18 |
Hb_004722_060 |
0.1398342993 |
- |
- |
PREDICTED: protein CLP1 homolog [Jatropha curcas] |
| 19 |
Hb_006400_040 |
0.1402373582 |
- |
- |
PREDICTED: uncharacterized protein LOC105642527 [Jatropha curcas] |
| 20 |
Hb_000732_260 |
0.1417184527 |
- |
- |
PREDICTED: telomerase Cajal body protein 1 [Jatropha curcas] |