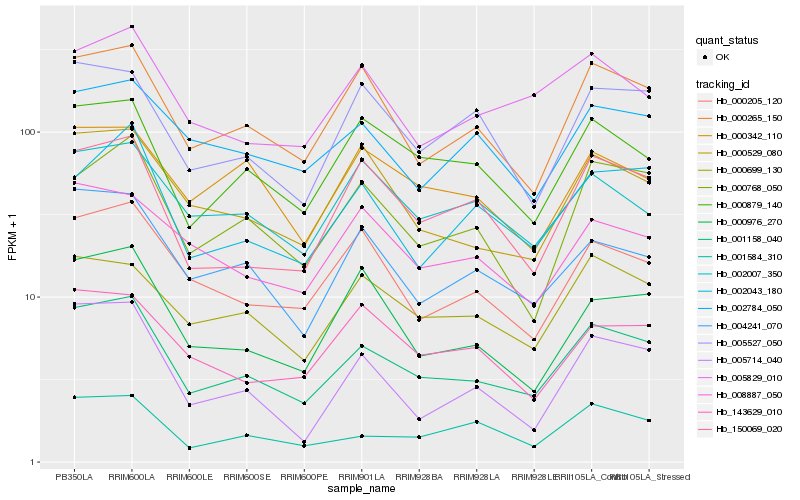
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001158_040 |
0.0 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |
| 2 |
Hb_000265_150 |
0.0864343535 |
- |
- |
PREDICTED: uncharacterized protein LOC105643541 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004241_070 |
0.098677625 |
- |
- |
PREDICTED: WD repeat-containing protein 25 [Jatropha curcas] |
| 4 |
Hb_000529_080 |
0.0986811334 |
- |
- |
PREDICTED: uncharacterized protein LOC105641675 [Jatropha curcas] |
| 5 |
Hb_000879_140 |
0.0999080409 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0283697-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_000699_130 |
0.1019113311 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_150069_020 |
0.1019893752 |
- |
- |
PREDICTED: ribosome biogenesis regulatory protein homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_000205_120 |
0.1027418877 |
- |
- |
PREDICTED: casein kinase I-like isoform X3 [Jatropha curcas] |
| 9 |
Hb_000768_050 |
0.1035491702 |
- |
- |
hypothetical protein RCOM_0845740 [Ricinus communis] |
| 10 |
Hb_001584_310 |
0.1062823995 |
- |
- |
hypothetical protein POPTR_0013s00630g [Populus trichocarpa] |
| 11 |
Hb_000976_270 |
0.1099288134 |
- |
- |
PREDICTED: uncharacterized protein LOC105646226 [Jatropha curcas] |
| 12 |
Hb_005527_050 |
0.1112824824 |
- |
- |
PREDICTED: uncharacterized protein LOC105643024 [Jatropha curcas] |
| 13 |
Hb_002043_180 |
0.1147316733 |
- |
- |
PREDICTED: NADP-specific glutamate dehydrogenase [Jatropha curcas] |
| 14 |
Hb_005829_010 |
0.1150205459 |
- |
- |
PREDICTED: uncharacterized protein LOC105640979 [Jatropha curcas] |
| 15 |
Hb_002784_050 |
0.1167687367 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6 [Jatropha curcas] |
| 16 |
Hb_008887_050 |
0.1169765033 |
- |
- |
PREDICTED: DNA-binding protein BIN4 [Jatropha curcas] |
| 17 |
Hb_002007_350 |
0.117824209 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 18 |
Hb_143629_010 |
0.1181455612 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 19 |
Hb_000342_110 |
0.1189040132 |
- |
- |
PREDICTED: uncharacterized protein LOC105638174 [Jatropha curcas] |
| 20 |
Hb_005714_040 |
0.1192251863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |