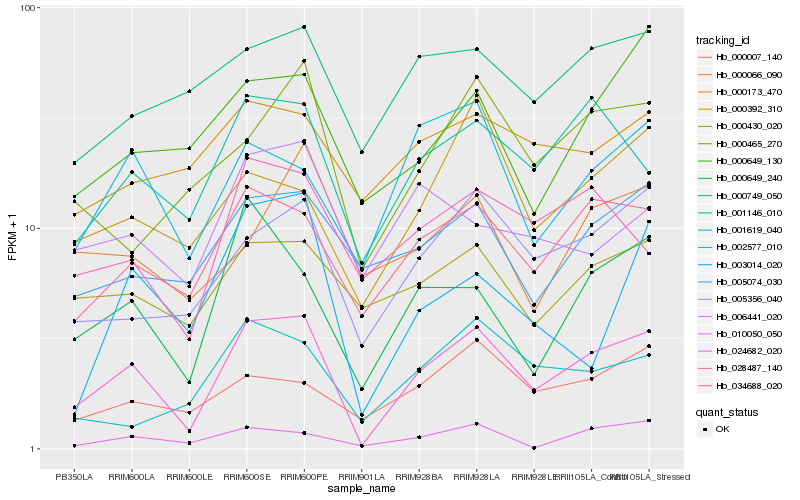
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024682_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s12710g [Populus trichocarpa] |
| 2 |
Hb_034688_020 |
0.1663617591 |
- |
- |
PREDICTED: uncharacterized protein LOC105642050 [Jatropha curcas] |
| 3 |
Hb_005356_040 |
0.1763902076 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 4 |
Hb_000649_240 |
0.1789911479 |
- |
- |
PREDICTED: probable peroxygenase 4 [Jatropha curcas] |
| 5 |
Hb_001146_010 |
0.1793464826 |
- |
- |
cystathionine gamma-synthase, putative [Ricinus communis] |
| 6 |
Hb_002577_010 |
0.1956903539 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 7 |
Hb_010050_050 |
0.1980135816 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog isoform X2 [Jatropha curcas] |
| 8 |
Hb_028487_140 |
0.199867774 |
- |
- |
PREDICTED: myosin-binding protein 2 [Jatropha curcas] |
| 9 |
Hb_005074_030 |
0.2006806971 |
- |
- |
- |
| 10 |
Hb_000430_020 |
0.2011487278 |
- |
- |
- |
| 11 |
Hb_000007_140 |
0.2015132404 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570 [Jatropha curcas] |
| 12 |
Hb_000465_270 |
0.2018933155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000392_310 |
0.2040877237 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001619_040 |
0.2050570414 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 15 |
Hb_000649_130 |
0.2081077088 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 16 |
Hb_000749_050 |
0.2082542122 |
- |
- |
mitochondrial thioredoxin [Hevea brasiliensis] |
| 17 |
Hb_003014_020 |
0.2126636815 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 18 |
Hb_006441_020 |
0.2177248566 |
- |
- |
PREDICTED: cadmium/zinc-transporting ATPase HMA3-like isoform X2 [Populus euphratica] |
| 19 |
Hb_000173_470 |
0.2193651658 |
- |
- |
PREDICTED: BSD domain-containing protein 1 [Jatropha curcas] |
| 20 |
Hb_000066_090 |
0.219772058 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |