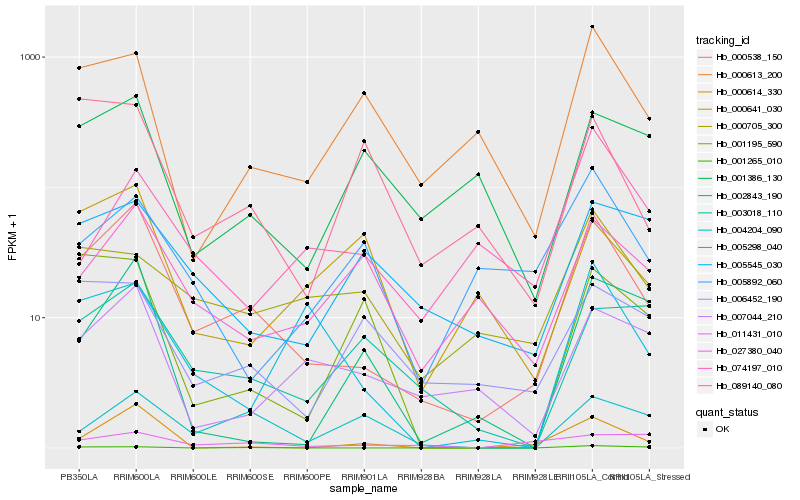
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000538_150 |
0.0 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 2 |
Hb_000614_330 |
0.2498260473 |
- |
- |
hypothetical protein POPTR_0008s18070g [Populus trichocarpa] |
| 3 |
Hb_007044_210 |
0.2680502438 |
transcription factor |
TF Family: MYB |
PREDICTED: protein rough sheath 2 homolog [Populus euphratica] |
| 4 |
Hb_027380_040 |
0.2765865084 |
- |
- |
hypothetical protein B456_011G046500, partial [Gossypium raimondii] |
| 5 |
Hb_000613_200 |
0.2784318428 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
acetyl-CoA C-acetyltransferase [Hevea brasiliensis] |
| 6 |
Hb_005298_040 |
0.2790862005 |
- |
- |
hypothetical protein CICLE_v10003181mg, partial [Citrus clementina] |
| 7 |
Hb_005545_030 |
0.2815685949 |
- |
- |
PREDICTED: uncharacterized protein LOC105645697 [Jatropha curcas] |
| 8 |
Hb_003018_110 |
0.2839145035 |
- |
- |
- |
| 9 |
Hb_000705_300 |
0.2845603502 |
- |
- |
PREDICTED: probable protein phosphatase 2C 24 [Jatropha curcas] |
| 10 |
Hb_006452_190 |
0.2858946874 |
- |
- |
PREDICTED: 3-isopropylmalate dehydrogenase 2, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_002843_190 |
0.294367664 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 7 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001265_010 |
0.2952167413 |
transcription factor |
TF Family: NAC |
NAC transcription factor 057 [Jatropha curcas] |
| 13 |
Hb_089140_080 |
0.2958621716 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105637962 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004204_090 |
0.2967601715 |
- |
- |
hypothetical protein JCGZ_11697 [Jatropha curcas] |
| 15 |
Hb_001386_130 |
0.2968746783 |
- |
- |
Mitochondrial import receptor subunit TOM20, putative [Ricinus communis] |
| 16 |
Hb_011431_010 |
0.2984217025 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 17 |
Hb_000641_030 |
0.299601612 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 18 |
Hb_001195_590 |
0.3042020061 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |
| 19 |
Hb_074197_010 |
0.3052075135 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 20 |
Hb_005892_060 |
0.3053795072 |
- |
- |
PREDICTED: uncharacterized protein LOC105632176 isoform X2 [Jatropha curcas] |