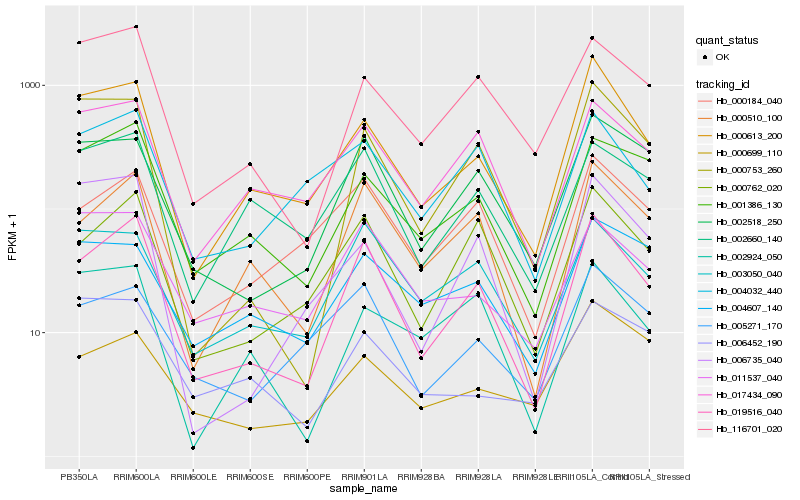
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000613_200 |
0.0 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
acetyl-CoA C-acetyltransferase [Hevea brasiliensis] |
| 2 |
Hb_019516_040 |
0.1386845441 |
desease resistance |
Gene Name: AAA |
Katanin p60 ATPase-containing subunit, putative [Ricinus communis] |
| 3 |
Hb_017434_090 |
0.1545422238 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000753_260 |
0.1575158107 |
transcription factor |
TF Family: MIKC |
MADS-box transcription factor 3 [Hevea brasiliensis] |
| 5 |
Hb_006735_040 |
0.1626752013 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_001386_130 |
0.1644687115 |
- |
- |
Mitochondrial import receptor subunit TOM20, putative [Ricinus communis] |
| 7 |
Hb_004607_140 |
0.1669979923 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor PLT1 [Populus euphratica] |
| 8 |
Hb_116701_020 |
0.1693297704 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000699_110 |
0.1696098584 |
- |
- |
hypothetical protein POPTR_0013s04180g [Populus trichocarpa] |
| 10 |
Hb_004032_440 |
0.17093525 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 1; Short=HMG-CoA reductase 1 [Hevea brasiliensis] |
| 11 |
Hb_002660_140 |
0.1733521626 |
- |
- |
hypothetical protein POPTR_0006s13060g [Populus trichocarpa] |
| 12 |
Hb_002518_250 |
0.1734049741 |
- |
- |
conserved hypothetical protein 10 [Hevea brasiliensis] |
| 13 |
Hb_005271_170 |
0.1750764019 |
- |
- |
PREDICTED: tetraspanin-10 isoform X3 [Vitis vinifera] |
| 14 |
Hb_000184_040 |
0.1756242078 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
PREDICTED: acetyl-CoA acetyltransferase, cytosolic 1-like isoform X2 [Nelumbo nucifera] |
| 15 |
Hb_000510_100 |
0.1760668902 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 16 |
Hb_003050_040 |
0.177775323 |
desease resistance |
Gene Name: AAA |
Katanin p60 ATPase-containing subunit, putative [Ricinus communis] |
| 17 |
Hb_006452_190 |
0.1785517505 |
- |
- |
PREDICTED: 3-isopropylmalate dehydrogenase 2, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_011537_040 |
0.1794048655 |
- |
- |
PREDICTED: uncharacterized protein LOC105646229 [Jatropha curcas] |
| 19 |
Hb_002924_050 |
0.1797323084 |
- |
- |
PREDICTED: uncharacterized protein LOC105629366 [Jatropha curcas] |
| 20 |
Hb_000762_020 |
0.1809812008 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |