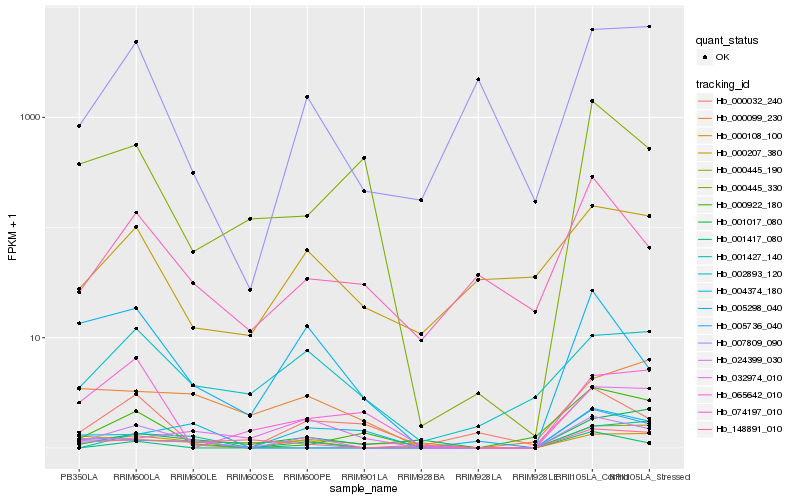
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024399_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005298_040 |
0.2763546594 |
- |
- |
hypothetical protein CICLE_v10003181mg, partial [Citrus clementina] |
| 3 |
Hb_001417_080 |
0.2806203785 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 4 |
Hb_000445_190 |
0.295605957 |
- |
- |
- |
| 5 |
Hb_002893_120 |
0.2962558108 |
- |
- |
PREDICTED: uncharacterized protein LOC105637692 [Jatropha curcas] |
| 6 |
Hb_007809_090 |
0.2976207808 |
- |
- |
protease inhibitor protein [Hevea brasiliensis] |
| 7 |
Hb_000108_100 |
0.2991607164 |
- |
- |
PREDICTED: DNA polymerase epsilon subunit 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_074197_010 |
0.3105757325 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 9 |
Hb_005736_040 |
0.313095388 |
- |
- |
- |
| 10 |
Hb_000032_240 |
0.3159552075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000922_180 |
0.3166544444 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO5 [Jatropha curcas] |
| 12 |
Hb_004374_180 |
0.3172768097 |
- |
- |
GMP synthase, putative [Ricinus communis] |
| 13 |
Hb_001017_080 |
0.3199650216 |
- |
- |
hypothetical protein POPTR_0010s06020g [Populus trichocarpa] |
| 14 |
Hb_148891_010 |
0.3202465144 |
- |
- |
- |
| 15 |
Hb_000207_380 |
0.32760015 |
- |
- |
sucrose transporter 6 [Hevea brasiliensis] |
| 16 |
Hb_065642_010 |
0.3320760392 |
- |
- |
- |
| 17 |
Hb_000099_230 |
0.3335521441 |
- |
- |
PREDICTED: F-box protein SKIP19-like [Gossypium raimondii] |
| 18 |
Hb_000445_330 |
0.3362103227 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_032974_010 |
0.3368349097 |
- |
- |
- |
| 20 |
Hb_001427_140 |
0.3401999575 |
- |
- |
PREDICTED: probable phospholipase A2 homolog 1 [Jatropha curcas] |