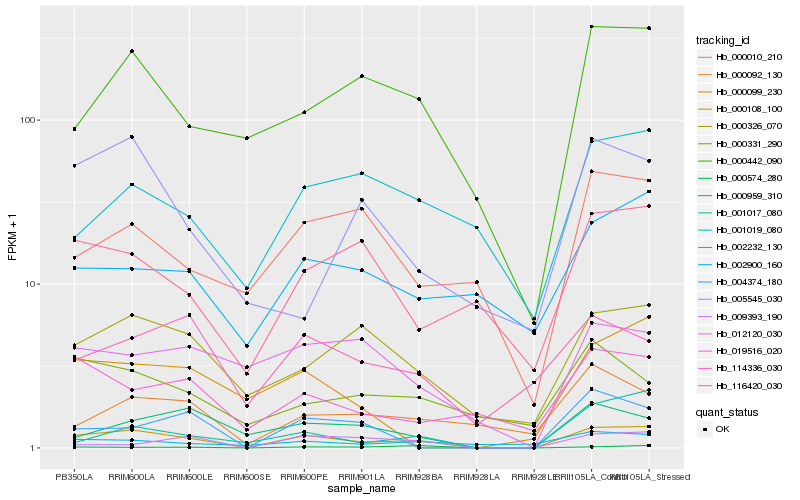
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000108_100 |
0.0 |
- |
- |
PREDICTED: DNA polymerase epsilon subunit 2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000326_070 |
0.2312973283 |
- |
- |
phenylalanyl-tRNA synthetase beta chain, putative [Ricinus communis] |
| 3 |
Hb_000442_090 |
0.2425593703 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like [Jatropha curcas] |
| 4 |
Hb_019516_020 |
0.255943373 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_116420_030 |
0.2565632493 |
- |
- |
hypothetical protein POPTR_0007s14760g [Populus trichocarpa] |
| 6 |
Hb_009393_190 |
0.2624309867 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase AIP2-like [Jatropha curcas] |
| 7 |
Hb_000092_130 |
0.264704806 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_000959_310 |
0.2657623725 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 9 |
Hb_000099_230 |
0.2658361771 |
- |
- |
PREDICTED: F-box protein SKIP19-like [Gossypium raimondii] |
| 10 |
Hb_004374_180 |
0.266120747 |
- |
- |
GMP synthase, putative [Ricinus communis] |
| 11 |
Hb_001017_080 |
0.2664114392 |
- |
- |
hypothetical protein POPTR_0010s06020g [Populus trichocarpa] |
| 12 |
Hb_000010_210 |
0.267444768 |
- |
- |
PREDICTED: centromere protein X [Jatropha curcas] |
| 13 |
Hb_012120_030 |
0.2683920915 |
- |
- |
- |
| 14 |
Hb_002232_130 |
0.2688142373 |
- |
- |
- |
| 15 |
Hb_114336_030 |
0.2695891958 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 16 |
Hb_002900_160 |
0.2707057955 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Jatropha curcas] |
| 17 |
Hb_000331_290 |
0.2711116459 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005545_030 |
0.2712698136 |
- |
- |
PREDICTED: uncharacterized protein LOC105645697 [Jatropha curcas] |
| 19 |
Hb_001019_080 |
0.271925008 |
- |
- |
hypothetical protein JCGZ_13280 [Jatropha curcas] |
| 20 |
Hb_000574_280 |
0.2720880102 |
- |
- |
hypothetical protein POPTR_0006s28780g [Populus trichocarpa] |