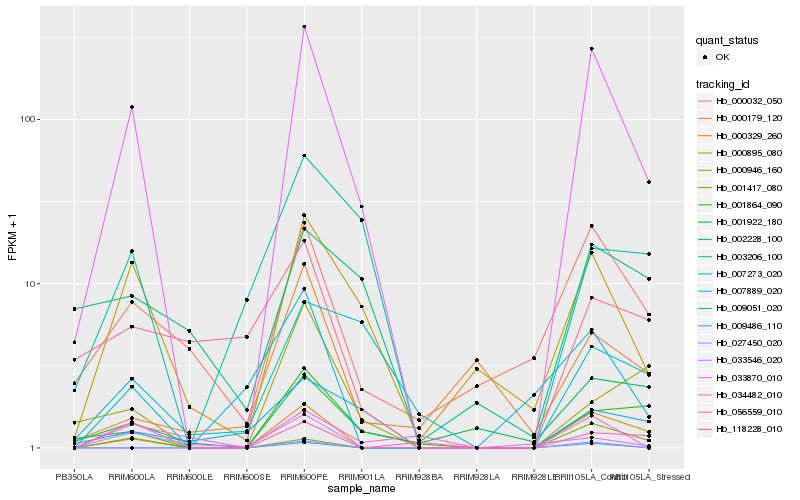
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033870_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104101109 isoform X1 [Nicotiana tomentosiformis] |
| 2 |
Hb_000329_260 |
0.2076721161 |
- |
- |
hypothetical protein MIMGU_mgv1a016162mg [Erythranthe guttata] |
| 3 |
Hb_000895_080 |
0.2310903166 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_000032_050 |
0.275275622 |
- |
- |
hypothetical protein RCOM_1691130 [Ricinus communis] |
| 5 |
Hb_001864_090 |
0.2893925969 |
- |
- |
- |
| 6 |
Hb_001417_080 |
0.3124315826 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 7 |
Hb_033546_020 |
0.3206686956 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor pho1, putative [Ricinus communis] |
| 8 |
Hb_007889_020 |
0.3217740064 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003206_100 |
0.3320881261 |
- |
- |
- |
| 10 |
Hb_056559_010 |
0.3333147414 |
- |
- |
PREDICTED: uncharacterized protein LOC104088595 [Nicotiana tomentosiformis] |
| 11 |
Hb_001922_180 |
0.3352420273 |
- |
- |
hevamine-A precursor, putative [Ricinus communis] |
| 12 |
Hb_009051_020 |
0.3410843567 |
- |
- |
Thioredoxin O1 [Morus notabilis] |
| 13 |
Hb_034482_010 |
0.342748571 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000946_160 |
0.3461928961 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 15 |
Hb_027450_020 |
0.3564429343 |
- |
- |
PREDICTED: B3 domain-containing protein At2g31420-like [Citrus sinensis] |
| 16 |
Hb_009486_110 |
0.3565723679 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 17 |
Hb_007273_020 |
0.3580655724 |
- |
- |
PREDICTED: probable U6 snRNA-associated Sm-like protein LSm4 [Cicer arietinum] |
| 18 |
Hb_002228_100 |
0.3598632609 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 19 |
Hb_118228_010 |
0.3677108183 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 20 |
Hb_000179_120 |
0.3698462856 |
- |
- |
PREDICTED: corepressor interacting with RBPJ 1-like [Jatropha curcas] |