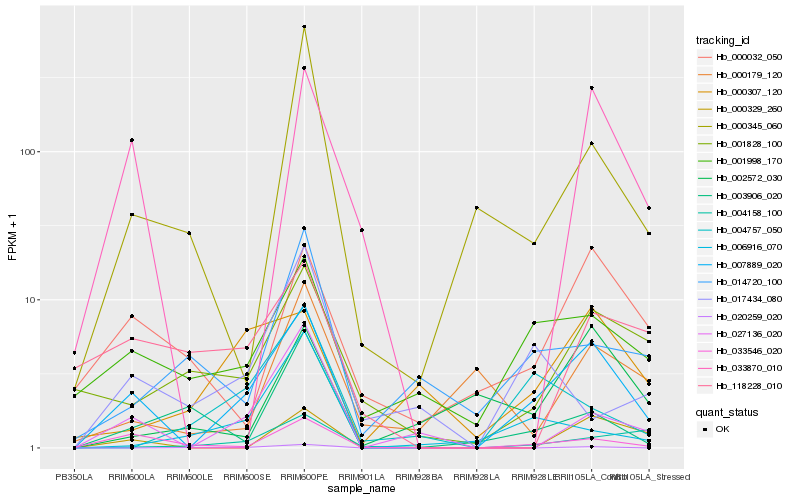
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007889_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_033546_020 |
0.2818185855 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor pho1, putative [Ricinus communis] |
| 3 |
Hb_001998_170 |
0.2925502355 |
- |
- |
BnaC07g08230D [Brassica napus] |
| 4 |
Hb_027136_020 |
0.3052145744 |
- |
- |
- |
| 5 |
Hb_000329_260 |
0.3199250803 |
- |
- |
hypothetical protein MIMGU_mgv1a016162mg [Erythranthe guttata] |
| 6 |
Hb_033870_010 |
0.3217740064 |
- |
- |
PREDICTED: uncharacterized protein LOC104101109 isoform X1 [Nicotiana tomentosiformis] |
| 7 |
Hb_000032_050 |
0.330157245 |
- |
- |
hypothetical protein RCOM_1691130 [Ricinus communis] |
| 8 |
Hb_001828_100 |
0.3324186203 |
- |
- |
PREDICTED: death-associated protein kinase dapk-1 [Jatropha curcas] |
| 9 |
Hb_000307_120 |
0.3463745554 |
- |
- |
hypothetical protein B456_002G120900 [Gossypium raimondii] |
| 10 |
Hb_000345_060 |
0.3464321535 |
- |
- |
PREDICTED: REF/SRPP-like protein At3g05500 [Jatropha curcas] |
| 11 |
Hb_004158_100 |
0.3541345581 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2 [Jatropha curcas] |
| 12 |
Hb_002572_030 |
0.3592242087 |
- |
- |
PREDICTED: diacylglycerol kinase theta [Jatropha curcas] |
| 13 |
Hb_020259_020 |
0.3622513269 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 4 [Jatropha curcas] |
| 14 |
Hb_003906_020 |
0.3635190953 |
- |
- |
PREDICTED: uncharacterized protein LOC105648535 [Jatropha curcas] |
| 15 |
Hb_006916_070 |
0.3636129325 |
transcription factor |
TF Family: MIKC |
PREDICTED: developmental protein SEPALLATA 1-like [Jatropha curcas] |
| 16 |
Hb_004757_050 |
0.3651304462 |
- |
- |
- |
| 17 |
Hb_014720_100 |
0.3664913051 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |
| 18 |
Hb_118228_010 |
0.3687227351 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 19 |
Hb_000179_120 |
0.3704873245 |
- |
- |
PREDICTED: corepressor interacting with RBPJ 1-like [Jatropha curcas] |
| 20 |
Hb_017434_080 |
0.3707808329 |
- |
- |
conserved hypothetical protein [Ricinus communis] |