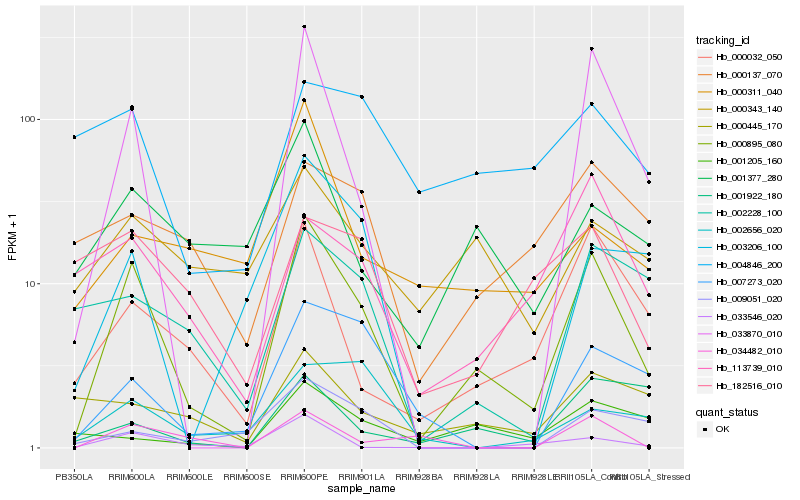
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000895_080 |
0.0 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1-like [Jatropha curcas] |
| 2 |
Hb_033870_010 |
0.2310903166 |
- |
- |
PREDICTED: uncharacterized protein LOC104101109 isoform X1 [Nicotiana tomentosiformis] |
| 3 |
Hb_000032_050 |
0.2687119221 |
- |
- |
hypothetical protein RCOM_1691130 [Ricinus communis] |
| 4 |
Hb_033546_020 |
0.2958878578 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor pho1, putative [Ricinus communis] |
| 5 |
Hb_007273_020 |
0.3097734823 |
- |
- |
PREDICTED: probable U6 snRNA-associated Sm-like protein LSm4 [Cicer arietinum] |
| 6 |
Hb_001377_280 |
0.3103878691 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Populus euphratica] |
| 7 |
Hb_113739_010 |
0.3119409651 |
- |
- |
hypothetical protein JCGZ_06379 [Jatropha curcas] |
| 8 |
Hb_002228_100 |
0.3135049843 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 9 |
Hb_003206_100 |
0.3148863177 |
- |
- |
- |
| 10 |
Hb_009051_020 |
0.3235186621 |
- |
- |
Thioredoxin O1 [Morus notabilis] |
| 11 |
Hb_000137_070 |
0.3269602319 |
- |
- |
PREDICTED: uncharacterized protein LOC105644263 isoform X1 [Jatropha curcas] |
| 12 |
Hb_034482_010 |
0.3277276642 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000343_140 |
0.3283588358 |
- |
- |
PREDICTED: uncharacterized protein LOC105634966 [Jatropha curcas] |
| 14 |
Hb_001922_180 |
0.3286487287 |
- |
- |
hevamine-A precursor, putative [Ricinus communis] |
| 15 |
Hb_000445_170 |
0.3291041229 |
- |
- |
PREDICTED: probable methyltransferase PMT10 [Jatropha curcas] |
| 16 |
Hb_001205_160 |
0.330338078 |
- |
- |
hypothetical protein POPTR_0005s12560g [Populus trichocarpa] |
| 17 |
Hb_000311_040 |
0.3352348299 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 18 |
Hb_182516_010 |
0.3409269256 |
- |
- |
PREDICTED: uncharacterized protein LOC105641595 [Jatropha curcas] |
| 19 |
Hb_004846_200 |
0.3422190184 |
- |
- |
PREDICTED: shikimate O-hydroxycinnamoyltransferase [Jatropha curcas] |
| 20 |
Hb_002656_020 |
0.3445861036 |
desease resistance |
Gene Name: NB-ARC |
ATP binding protein, putative [Ricinus communis] |