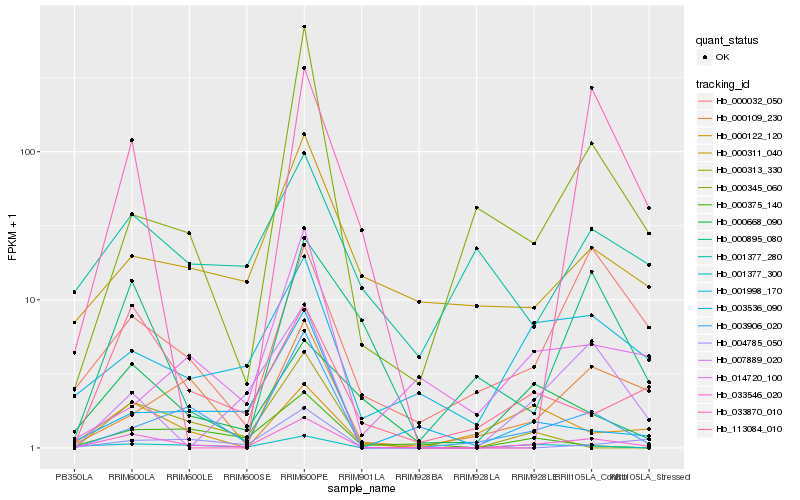
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033546_020 |
0.0 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor pho1, putative [Ricinus communis] |
| 2 |
Hb_001377_300 |
0.2612043259 |
- |
- |
flavonoid 3-hydroxylase, putative [Ricinus communis] |
| 3 |
Hb_001998_170 |
0.2704226948 |
- |
- |
BnaC07g08230D [Brassica napus] |
| 4 |
Hb_113084_010 |
0.2712209159 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 5 |
Hb_003906_020 |
0.281668932 |
- |
- |
PREDICTED: uncharacterized protein LOC105648535 [Jatropha curcas] |
| 6 |
Hb_007889_020 |
0.2818185855 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000311_040 |
0.2873200261 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 8 |
Hb_000668_090 |
0.2873211745 |
- |
- |
senescence-associated family protein [Populus trichocarpa] |
| 9 |
Hb_000032_050 |
0.2954691099 |
- |
- |
hypothetical protein RCOM_1691130 [Ricinus communis] |
| 10 |
Hb_000895_080 |
0.2958878578 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1-like [Jatropha curcas] |
| 11 |
Hb_000345_060 |
0.3014958068 |
- |
- |
PREDICTED: REF/SRPP-like protein At3g05500 [Jatropha curcas] |
| 12 |
Hb_003536_090 |
0.3026643588 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 13 |
Hb_000109_230 |
0.3098084991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001377_280 |
0.3120755499 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Populus euphratica] |
| 15 |
Hb_000375_140 |
0.3133316194 |
- |
- |
PREDICTED: monoacylglycerol lipase ABHD6 [Jatropha curcas] |
| 16 |
Hb_000313_330 |
0.3136480825 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase [Jatropha curcas] |
| 17 |
Hb_004785_050 |
0.3172789946 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 18 |
Hb_033870_010 |
0.3206686956 |
- |
- |
PREDICTED: uncharacterized protein LOC104101109 isoform X1 [Nicotiana tomentosiformis] |
| 19 |
Hb_000122_120 |
0.3230999222 |
- |
- |
PREDICTED: arogenate dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_014720_100 |
0.3232273854 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |