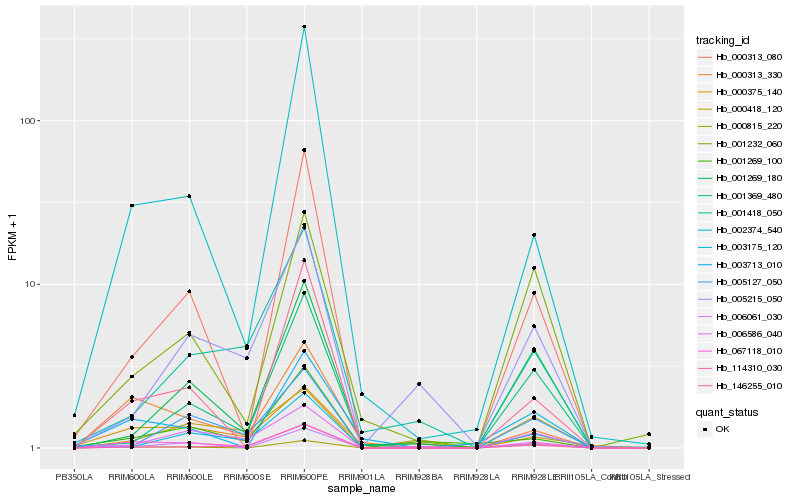
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_067118_010 |
0.0 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 2 |
Hb_000313_330 |
0.1505234069 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase [Jatropha curcas] |
| 3 |
Hb_000375_140 |
0.2137301199 |
- |
- |
PREDICTED: monoacylglycerol lipase ABHD6 [Jatropha curcas] |
| 4 |
Hb_006061_030 |
0.2139980783 |
- |
- |
PREDICTED: fe(2+) transport protein 1-like [Jatropha curcas] |
| 5 |
Hb_003713_010 |
0.2190218661 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_146255_010 |
0.2224442173 |
- |
- |
PREDICTED: uncharacterized protein LOC105643649 [Jatropha curcas] |
| 7 |
Hb_002374_540 |
0.2254592035 |
- |
- |
RecName: Full=Esterase; AltName: Full=Early nodule-specific protein homolog; AltName: Full=Latex allergen Hev b 13; AltName: Allergen=Hev b 13; Flags: Precursor [Hevea brasiliensis] |
| 8 |
Hb_000313_080 |
0.2256939181 |
rubber biosynthesis |
Gene Name: CPT3 |
cis-prenyltransferase 3 [Hevea brasiliensis] |
| 9 |
Hb_000815_220 |
0.2267106384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006586_040 |
0.2318664076 |
- |
- |
hypothetical protein POPTR_0011s11750g [Populus trichocarpa] |
| 11 |
Hb_001232_060 |
0.234588028 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001269_180 |
0.2391716694 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_003175_120 |
0.2487474129 |
- |
- |
PREDICTED: uncharacterized protein LOC105141472 isoform X1 [Populus euphratica] |
| 14 |
Hb_001269_100 |
0.2491446019 |
- |
- |
plasma membrane ATPase 1, partial [Triticum aestivum] |
| 15 |
Hb_005127_050 |
0.2549387702 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA2-like [Populus euphratica] |
| 16 |
Hb_114310_030 |
0.2558514787 |
- |
- |
JHL18I08.8 [Jatropha curcas] |
| 17 |
Hb_001369_480 |
0.256745122 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0012s14660g [Populus trichocarpa] |
| 18 |
Hb_001418_050 |
0.2584460744 |
- |
- |
PREDICTED: uncharacterized protein LOC105631088 [Jatropha curcas] |
| 19 |
Hb_000418_120 |
0.2587961692 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase [Jatropha curcas] |
| 20 |
Hb_005215_050 |
0.2591045369 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g37250 [Jatropha curcas] |