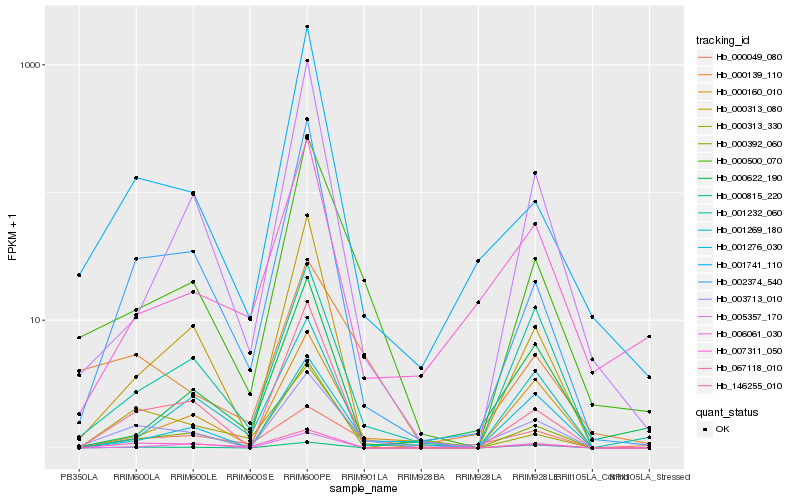
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003713_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000500_070 |
0.2096540343 |
- |
- |
hypothetical protein POPTR_0015s12340g [Populus trichocarpa] |
| 3 |
Hb_001232_060 |
0.2102776621 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000815_220 |
0.2141796025 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_067118_010 |
0.2190218661 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 6 |
Hb_000160_010 |
0.2216225346 |
- |
- |
PREDICTED: uncharacterized protein LOC105638816 [Jatropha curcas] |
| 7 |
Hb_000139_110 |
0.2243100083 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 8 |
Hb_146255_010 |
0.224650837 |
- |
- |
PREDICTED: uncharacterized protein LOC105643649 [Jatropha curcas] |
| 9 |
Hb_000313_080 |
0.2257365779 |
rubber biosynthesis |
Gene Name: CPT3 |
cis-prenyltransferase 3 [Hevea brasiliensis] |
| 10 |
Hb_000049_080 |
0.2306932164 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 11 |
Hb_001276_030 |
0.2320676297 |
- |
- |
PREDICTED: uncharacterized protein LOC104881333 [Vitis vinifera] |
| 12 |
Hb_002374_540 |
0.2328968083 |
- |
- |
RecName: Full=Esterase; AltName: Full=Early nodule-specific protein homolog; AltName: Full=Latex allergen Hev b 13; AltName: Allergen=Hev b 13; Flags: Precursor [Hevea brasiliensis] |
| 13 |
Hb_000313_330 |
0.2331793017 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase [Jatropha curcas] |
| 14 |
Hb_006061_030 |
0.233445995 |
- |
- |
PREDICTED: fe(2+) transport protein 1-like [Jatropha curcas] |
| 15 |
Hb_001741_110 |
0.2394831626 |
rubber biosynthesis |
Gene Name: REF7 |
REF/SRPP-like protein [Glycine soja] |
| 16 |
Hb_007311_050 |
0.2469937545 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000392_060 |
0.2498228463 |
- |
- |
hypothetical protein JCGZ_11629 [Jatropha curcas] |
| 18 |
Hb_001269_180 |
0.2613836436 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_005357_170 |
0.2616984928 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 20 |
Hb_000622_190 |
0.2651477368 |
- |
- |
PREDICTED: uncharacterized protein LOC105642890 [Jatropha curcas] |