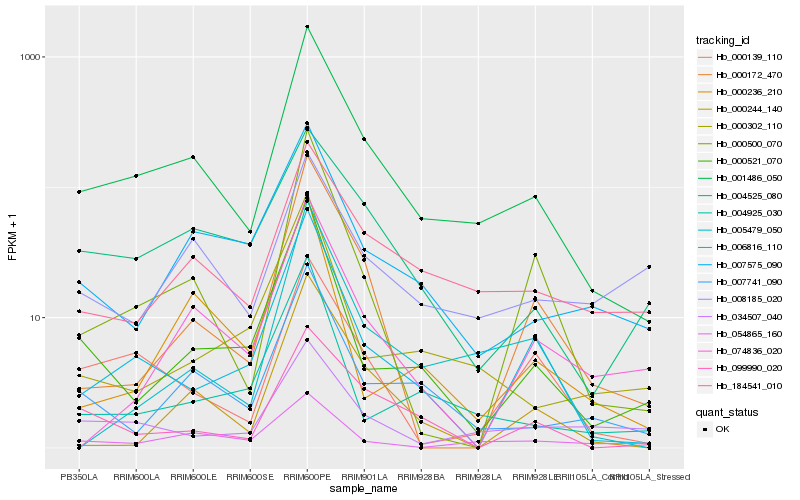
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001486_050 |
0.0 |
- |
- |
PREDICTED: arabinogalactan peptide 13-like [Jatropha curcas] |
| 2 |
Hb_184541_010 |
0.1681600921 |
- |
- |
PREDICTED: selT-like protein [Jatropha curcas] |
| 3 |
Hb_007741_090 |
0.1725049512 |
- |
- |
Autophagy-related 8f -like protein [Gossypium arboreum] |
| 4 |
Hb_004525_080 |
0.1830406524 |
- |
- |
Sugar transporter ERD6-like 6 [Morus notabilis] |
| 5 |
Hb_007575_090 |
0.1844730591 |
- |
- |
putative. proteolipid subunit of vacuolar H+ ATPase, partial [Zea mays] |
| 6 |
Hb_000139_110 |
0.1854529374 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 7 |
Hb_006816_110 |
0.1873448138 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 1 [Jatropha curcas] |
| 8 |
Hb_000521_070 |
0.1963202066 |
- |
- |
PREDICTED: uncharacterized protein LOC105650323 [Jatropha curcas] |
| 9 |
Hb_000500_070 |
0.1989159906 |
- |
- |
hypothetical protein POPTR_0015s12340g [Populus trichocarpa] |
| 10 |
Hb_099990_020 |
0.2045645628 |
- |
- |
hypothetical protein POPTR_0006s28940g [Populus trichocarpa] |
| 11 |
Hb_054865_160 |
0.2079054864 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 12 |
Hb_005479_050 |
0.2080572422 |
- |
- |
- |
| 13 |
Hb_034507_040 |
0.2106642878 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 14 |
Hb_004925_030 |
0.2113818356 |
- |
- |
EXORDIUM like 1 [Theobroma cacao] |
| 15 |
Hb_000302_110 |
0.2142641342 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000172_470 |
0.2157355387 |
- |
- |
hypothetical protein POPTR_0005s08310g [Populus trichocarpa] |
| 17 |
Hb_000244_140 |
0.2186564122 |
- |
- |
PREDICTED: cell wall / vacuolar inhibitor of fructosidase 2 [Jatropha curcas] |
| 18 |
Hb_008185_020 |
0.2207189709 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 19 |
Hb_074836_020 |
0.2264614707 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_000236_210 |
0.2269538916 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |