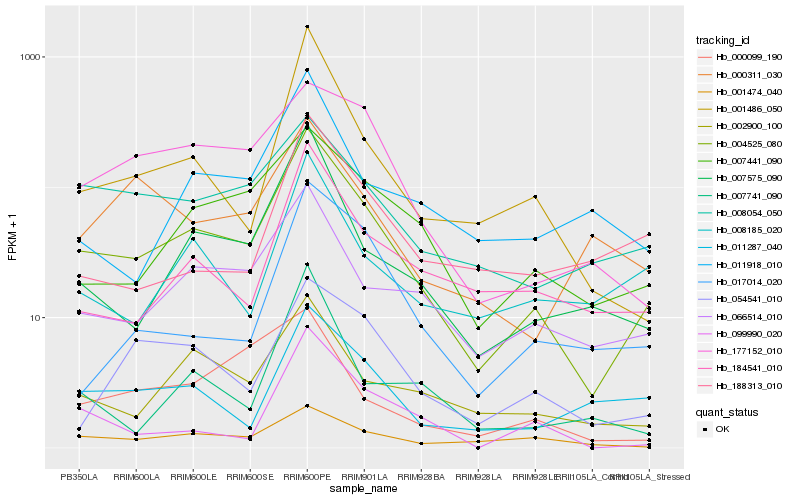
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004525_080 |
0.0 |
- |
- |
Sugar transporter ERD6-like 6 [Morus notabilis] |
| 2 |
Hb_066514_010 |
0.1699767682 |
- |
- |
hypothetical protein RCOM_0204720 [Ricinus communis] |
| 3 |
Hb_007575_090 |
0.1711322733 |
- |
- |
putative. proteolipid subunit of vacuolar H+ ATPase, partial [Zea mays] |
| 4 |
Hb_002900_100 |
0.1815139035 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 5 |
Hb_001486_050 |
0.1830406524 |
- |
- |
PREDICTED: arabinogalactan peptide 13-like [Jatropha curcas] |
| 6 |
Hb_007441_090 |
0.1855784704 |
- |
- |
nucleoside diphosphate kinase 1 [Hevea brasiliensis] |
| 7 |
Hb_177152_010 |
0.1960650353 |
- |
- |
glycosyl hydrolase family 38 family protein [Populus trichocarpa] |
| 8 |
Hb_008054_050 |
0.1964007532 |
- |
- |
PREDICTED: uncharacterized protein LOC101212042 [Cucumis sativus] |
| 9 |
Hb_184541_010 |
0.1975387309 |
- |
- |
PREDICTED: selT-like protein [Jatropha curcas] |
| 10 |
Hb_011287_040 |
0.2024640961 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001474_040 |
0.2039768158 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 12 |
Hb_008185_020 |
0.208286292 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 13 |
Hb_054541_010 |
0.209502452 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 14 |
Hb_011918_010 |
0.2123965631 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 15 |
Hb_099990_020 |
0.2192283314 |
- |
- |
hypothetical protein POPTR_0006s28940g [Populus trichocarpa] |
| 16 |
Hb_007741_090 |
0.2228611186 |
- |
- |
Autophagy-related 8f -like protein [Gossypium arboreum] |
| 17 |
Hb_000311_030 |
0.2230272209 |
- |
- |
hypothetical protein B456_002G227600 [Gossypium raimondii] |
| 18 |
Hb_017014_020 |
0.2239974015 |
- |
- |
- |
| 19 |
Hb_000099_190 |
0.2240643745 |
- |
- |
PREDICTED: putative cell division cycle ATPase [Jatropha curcas] |
| 20 |
Hb_188313_010 |
0.2264442646 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |