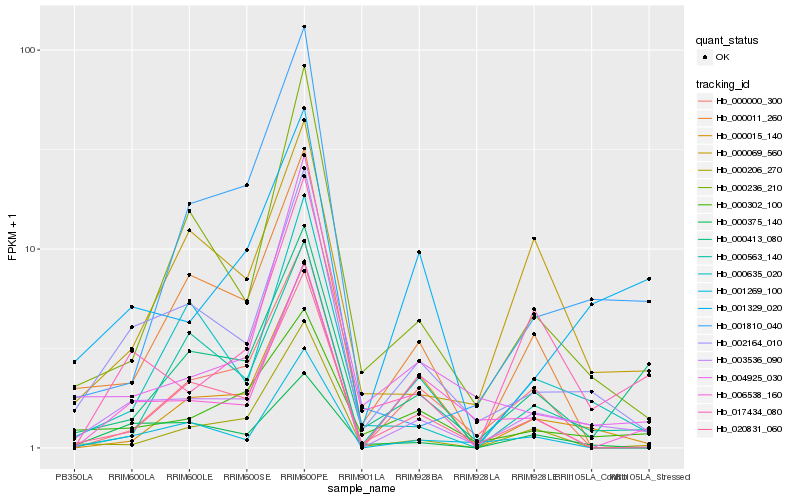
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003536_090 |
0.0 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 2 |
Hb_002164_010 |
0.1220566853 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 3 |
Hb_000236_210 |
0.1880446818 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 4 |
Hb_017434_080 |
0.1925251963 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_020831_060 |
0.1929465852 |
- |
- |
PREDICTED: uncharacterized protein LOC105631088 [Jatropha curcas] |
| 6 |
Hb_001269_100 |
0.1955154282 |
- |
- |
plasma membrane ATPase 1, partial [Triticum aestivum] |
| 7 |
Hb_000302_100 |
0.1965855558 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 8 |
Hb_000206_270 |
0.2013015235 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_000635_020 |
0.2026542885 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000413_080 |
0.202717081 |
- |
- |
- |
| 11 |
Hb_004925_030 |
0.2073469 |
- |
- |
EXORDIUM like 1 [Theobroma cacao] |
| 12 |
Hb_000375_140 |
0.2073965147 |
- |
- |
PREDICTED: monoacylglycerol lipase ABHD6 [Jatropha curcas] |
| 13 |
Hb_001329_020 |
0.210034837 |
- |
- |
PREDICTED: probable boron transporter 2 [Populus euphratica] |
| 14 |
Hb_006538_160 |
0.2112467975 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001810_040 |
0.2123952118 |
- |
- |
- |
| 16 |
Hb_000011_260 |
0.2138303574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000015_140 |
0.2153628716 |
- |
- |
PREDICTED: CASP-like protein 4C1 [Vitis vinifera] |
| 18 |
Hb_000563_140 |
0.2182039393 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 19 |
Hb_000069_560 |
0.2186232654 |
- |
- |
hypothetical protein POPTR_0002s16980g [Populus trichocarpa] |
| 20 |
Hb_000000_300 |
0.221568331 |
- |
- |
PREDICTED: pathogenesis-related protein 5 [Jatropha curcas] |