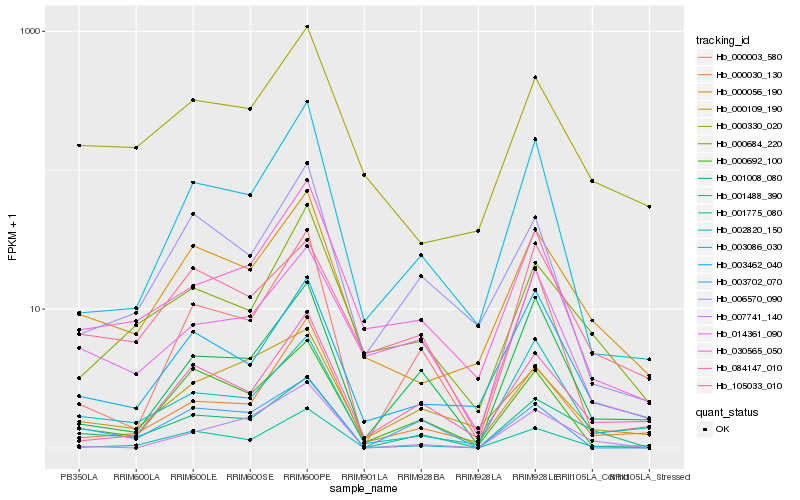
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001488_390 |
0.0 |
- |
- |
SP1L, putative [Ricinus communis] |
| 2 |
Hb_000056_190 |
0.1842003637 |
- |
- |
hypothetical protein [Ricinus communis] |
| 3 |
Hb_000684_220 |
0.1927391766 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_000692_100 |
0.1997965545 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 5 |
Hb_003462_040 |
0.2000148216 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_105033_010 |
0.2001520544 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001008_080 |
0.20250394 |
- |
- |
PREDICTED: acyl-protein thioesterase 2 [Jatropha curcas] |
| 8 |
Hb_002820_150 |
0.2030845381 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_003702_070 |
0.2037519353 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_000003_580 |
0.209225747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_014361_090 |
0.2093820476 |
- |
- |
acyl-CoA binding protein 3A [Vernicia fordii] |
| 12 |
Hb_003086_030 |
0.2105981227 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 13 |
Hb_000109_190 |
0.2124579482 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 14 |
Hb_030565_050 |
0.2129208581 |
- |
- |
UDP-D-GLUCURONATE 4-EPIMERASE 6 family protein [Populus trichocarpa] |
| 15 |
Hb_001775_080 |
0.2162904941 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 16 |
Hb_000330_020 |
0.2177123759 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 17 |
Hb_084147_010 |
0.2189917713 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_006570_090 |
0.2201054425 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 19 |
Hb_000030_130 |
0.2229697567 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_007741_140 |
0.2249945651 |
- |
- |
PREDICTED: SPX domain-containing protein 3-like isoform X2 [Populus euphratica] |