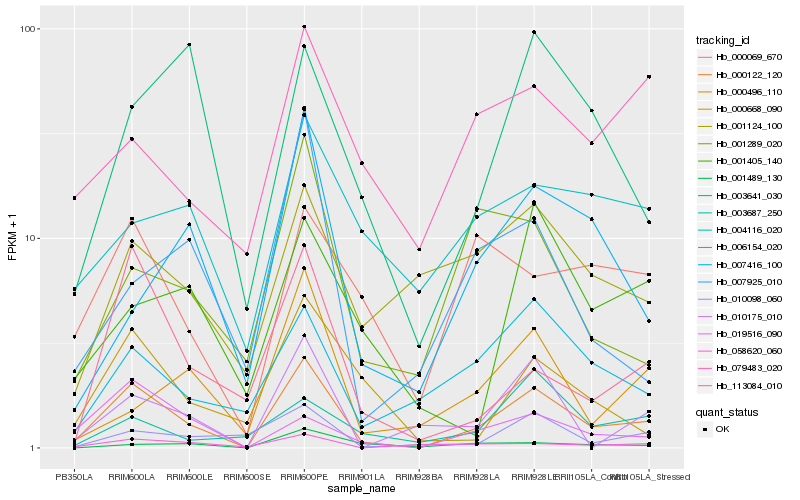
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000122_120 |
0.0 |
- |
- |
PREDICTED: arogenate dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_010175_010 |
0.2619388559 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006154_020 |
0.2620736294 |
transcription factor |
TF Family: C2H2 |
hypothetical protein RCOM_1621120 [Ricinus communis] |
| 4 |
Hb_113084_010 |
0.2636455331 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 5 |
Hb_000668_090 |
0.2703650644 |
- |
- |
senescence-associated family protein [Populus trichocarpa] |
| 6 |
Hb_058620_060 |
0.2739801446 |
- |
- |
uncharacterized protein LOC105646159 precursor [Jatropha curcas] |
| 7 |
Hb_019516_090 |
0.2742186941 |
- |
- |
hypothetical protein POPTR_0004s06350g [Populus trichocarpa] |
| 8 |
Hb_010098_060 |
0.2746125493 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g17580 [Jatropha curcas] |
| 9 |
Hb_000069_670 |
0.2795612317 |
- |
- |
PREDICTED: uncharacterized protein LOC105642887 [Jatropha curcas] |
| 10 |
Hb_001124_100 |
0.2804059069 |
- |
- |
PREDICTED: vesicle-associated protein 2-2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000496_110 |
0.2845900832 |
- |
- |
PREDICTED: uncharacterized protein LOC105650214 [Jatropha curcas] |
| 12 |
Hb_001289_020 |
0.2865651172 |
- |
- |
PREDICTED: uncharacterized protein LOC105635827 [Jatropha curcas] |
| 13 |
Hb_007416_100 |
0.2913776729 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 5 [Jatropha curcas] |
| 14 |
Hb_003641_030 |
0.2918457506 |
- |
- |
PREDICTED: uncharacterized protein LOC105638015 [Jatropha curcas] |
| 15 |
Hb_001489_130 |
0.2923614989 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |
| 16 |
Hb_079483_020 |
0.2936010647 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_004116_020 |
0.2948276438 |
- |
- |
fad NAD binding oxidoreductases, putative [Ricinus communis] |
| 18 |
Hb_001405_140 |
0.2964765551 |
- |
- |
hypothetical protein POPTR_0004s08350g, partial [Populus trichocarpa] |
| 19 |
Hb_007925_010 |
0.2989411482 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 1 [Jatropha curcas] |
| 20 |
Hb_003687_250 |
0.3003780979 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 1-like [Jatropha curcas] |