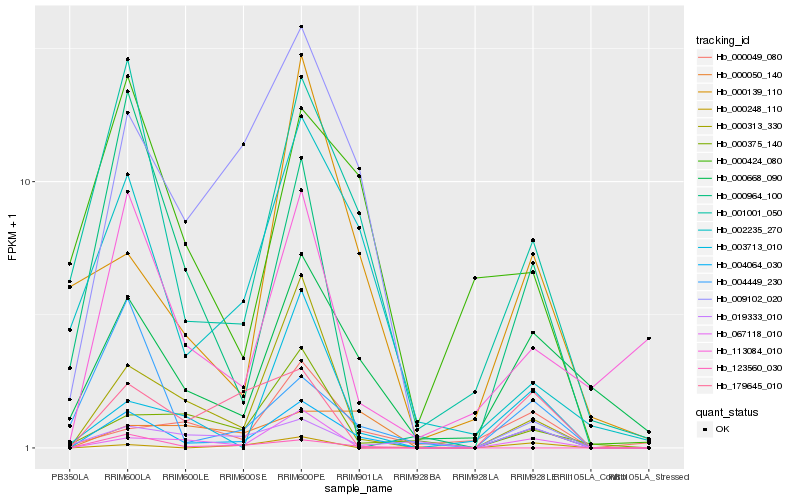
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004064_030 |
0.0 |
- |
- |
aldose-1-epimerase, putative [Ricinus communis] |
| 2 |
Hb_001001_050 |
0.2239197091 |
- |
- |
CXE protein [Hevea brasiliensis] |
| 3 |
Hb_000668_090 |
0.2506409436 |
- |
- |
senescence-associated family protein [Populus trichocarpa] |
| 4 |
Hb_000313_330 |
0.2705906831 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase [Jatropha curcas] |
| 5 |
Hb_002235_270 |
0.2888217016 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 6 |
Hb_067118_010 |
0.301839128 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 7 |
Hb_179645_010 |
0.3021877615 |
- |
- |
- |
| 8 |
Hb_113084_010 |
0.3055243773 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 9 |
Hb_019333_010 |
0.3058933358 |
- |
- |
PREDICTED: uncharacterized protein LOC105633210 [Jatropha curcas] |
| 10 |
Hb_003713_010 |
0.3118213123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004449_230 |
0.3130971553 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000050_140 |
0.3156301308 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2 isoform X1 [Populus euphratica] |
| 13 |
Hb_000424_080 |
0.317462475 |
- |
- |
PREDICTED: endoglucanase 16 [Jatropha curcas] |
| 14 |
Hb_000964_100 |
0.3186831528 |
- |
- |
PREDICTED: uncharacterized protein LOC105628699 [Jatropha curcas] |
| 15 |
Hb_000049_080 |
0.3241780072 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 16 |
Hb_009102_020 |
0.32662432 |
- |
- |
hypothetical protein EUGRSUZ_I01262 [Eucalyptus grandis] |
| 17 |
Hb_000139_110 |
0.3297731112 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 18 |
Hb_123560_030 |
0.3309316954 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000375_140 |
0.3317886215 |
- |
- |
PREDICTED: monoacylglycerol lipase ABHD6 [Jatropha curcas] |
| 20 |
Hb_000248_110 |
0.338716318 |
- |
- |
hypothetical protein VITISV_037580 [Vitis vinifera] |