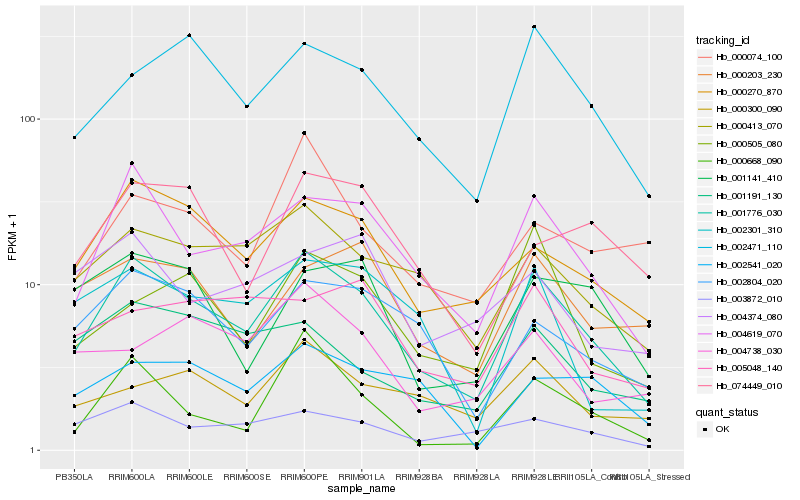
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001776_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004619_070 |
0.1165104936 |
- |
- |
hypothetical protein POPTR_0006s00550g [Populus trichocarpa] |
| 3 |
Hb_000270_870 |
0.1429459332 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 4 |
Hb_000413_070 |
0.1526617257 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002471_110 |
0.1590750863 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |
| 6 |
Hb_002804_020 |
0.1678122312 |
- |
- |
epsin N-terminal homology domain-containing family protein [Populus trichocarpa] |
| 7 |
Hb_003872_010 |
0.1704102155 |
- |
- |
hypothetical protein PRUPE_ppa013215mg [Prunus persica] |
| 8 |
Hb_000300_090 |
0.1807464326 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_000668_090 |
0.1824391831 |
- |
- |
senescence-associated family protein [Populus trichocarpa] |
| 10 |
Hb_002301_310 |
0.1876619889 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_074449_010 |
0.1897507322 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 12 |
Hb_000203_230 |
0.1903131623 |
- |
- |
PREDICTED: probable inactive dual specificity protein phosphatase-like At4g18593 [Jatropha curcas] |
| 13 |
Hb_001191_130 |
0.1903347214 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g71810, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002541_020 |
0.1905875913 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 15 |
Hb_000505_080 |
0.1905903613 |
- |
- |
Inner membrane transport protein yjjL, putative [Ricinus communis] |
| 16 |
Hb_001141_410 |
0.1926709033 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 17 |
Hb_000074_100 |
0.1939001129 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004738_030 |
0.2005061323 |
- |
- |
Inactive protein kinase [Gossypium arboreum] |
| 19 |
Hb_005048_140 |
0.200698908 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004374_080 |
0.2035059988 |
- |
- |
PREDICTED: uncharacterized protein LOC105645766 [Jatropha curcas] |