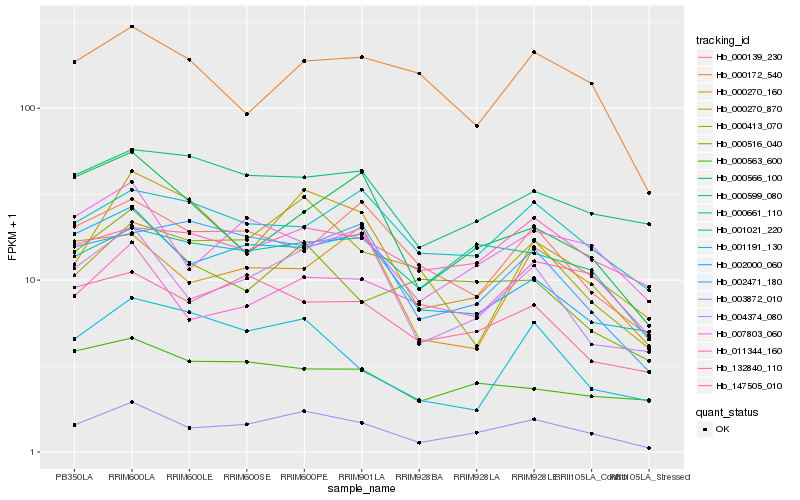
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003872_010 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa013215mg [Prunus persica] |
| 2 |
Hb_004374_080 |
0.1282228207 |
- |
- |
PREDICTED: uncharacterized protein LOC105645766 [Jatropha curcas] |
| 3 |
Hb_007803_060 |
0.135447581 |
- |
- |
PREDICTED: uncharacterized protein LOC105632794 [Jatropha curcas] |
| 4 |
Hb_000270_870 |
0.1400584571 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 5 |
Hb_002000_060 |
0.142000505 |
- |
- |
PREDICTED: protein LIKE COV 2 [Jatropha curcas] |
| 6 |
Hb_000661_110 |
0.1420327362 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 7 |
Hb_147505_010 |
0.1465652668 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 8 |
Hb_000516_040 |
0.1572721812 |
- |
- |
PREDICTED: uncharacterized protein LOC105649502 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001021_220 |
0.1576260566 |
- |
- |
PREDICTED: spermatogenesis-associated protein 20 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002471_180 |
0.15784105 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 11 |
Hb_000599_080 |
0.1603725821 |
- |
- |
PREDICTED: TOM1-like protein 1 [Malus domestica] |
| 12 |
Hb_000172_540 |
0.1604258539 |
- |
- |
hypothetical protein POPTR_0005s08350g [Populus trichocarpa] |
| 13 |
Hb_000566_100 |
0.1606155058 |
- |
- |
hypothetical protein JCGZ_17993 [Jatropha curcas] |
| 14 |
Hb_000139_230 |
0.1608887125 |
transcription factor |
TF Family: SNF2 |
PREDICTED: probable ATP-dependent DNA helicase CHR12 [Jatropha curcas] |
| 15 |
Hb_000413_070 |
0.1630142324 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000270_160 |
0.163688614 |
- |
- |
PREDICTED: uncharacterized protein LOC102625351 [Citrus sinensis] |
| 17 |
Hb_001191_130 |
0.1641777692 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g71810, chloroplastic [Jatropha curcas] |
| 18 |
Hb_132840_110 |
0.1643612156 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_000563_600 |
0.1649472401 |
- |
- |
PREDICTED: uncharacterized protein LOC104428079 [Eucalyptus grandis] |
| 20 |
Hb_011344_160 |
0.1655477298 |
- |
- |
PREDICTED: primary amine oxidase [Jatropha curcas] |