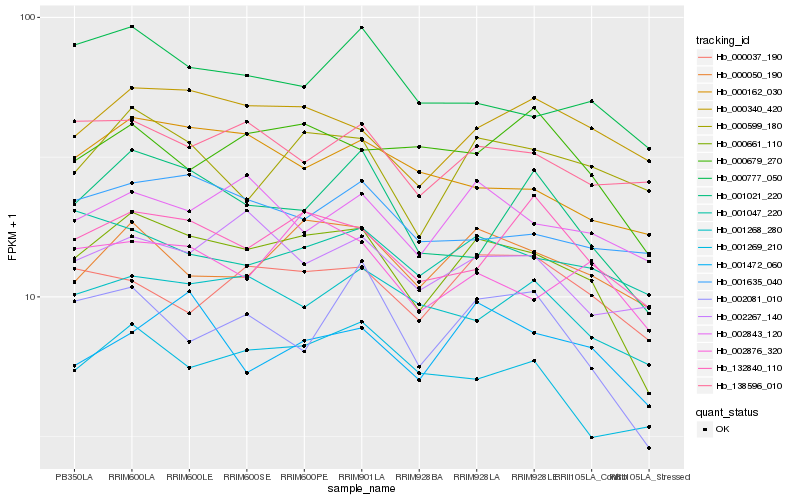
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000661_110 |
0.0 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 2 |
Hb_000050_190 |
0.0981098275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_132840_110 |
0.0987177136 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_002876_320 |
0.1008535592 |
- |
- |
PREDICTED: uncharacterized protein LOC105632052 [Jatropha curcas] |
| 5 |
Hb_001021_220 |
0.1011168437 |
- |
- |
PREDICTED: spermatogenesis-associated protein 20 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001268_280 |
0.1018799504 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002081_010 |
0.1035813173 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001269_210 |
0.1043004602 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g76280 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000340_420 |
0.1048484156 |
- |
- |
PREDICTED: LEC14B protein isoform X2 [Jatropha curcas] |
| 10 |
Hb_002843_120 |
0.1049909657 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000162_030 |
0.1052110147 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_000679_270 |
0.1057955293 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 13 |
Hb_001472_060 |
0.1061683364 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000599_180 |
0.106552273 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_000777_050 |
0.107801694 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 16 |
Hb_002267_140 |
0.1088780105 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 17 |
Hb_001047_220 |
0.1097120954 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 18 |
Hb_000037_190 |
0.1100621174 |
- |
- |
PREDICTED: uncharacterized protein LOC105641500 [Jatropha curcas] |
| 19 |
Hb_001635_040 |
0.1115837547 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 20 |
Hb_138596_010 |
0.1118690424 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |