| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
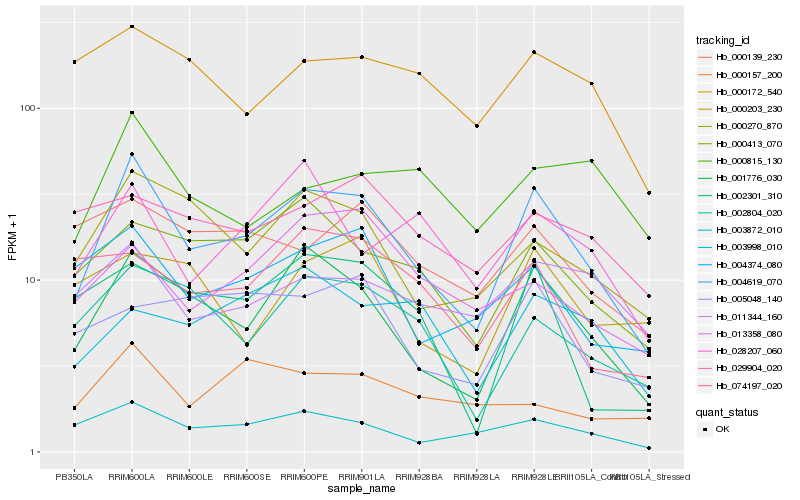
Hb_004619_070 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s00550g [Populus trichocarpa] |
| 2 |
Hb_001776_030 |
0.1165104936 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003872_010 |
0.1723503227 |
- |
- |
hypothetical protein PRUPE_ppa013215mg [Prunus persica] |
| 4 |
Hb_000270_870 |
0.1725867733 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 5 |
Hb_000413_070 |
0.1798482509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004374_080 |
0.1809685718 |
- |
- |
PREDICTED: uncharacterized protein LOC105645766 [Jatropha curcas] |
| 7 |
Hb_002804_020 |
0.1832617964 |
- |
- |
epsin N-terminal homology domain-containing family protein [Populus trichocarpa] |
| 8 |
Hb_002301_310 |
0.1959152496 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_028207_060 |
0.2038886478 |
- |
- |
PREDICTED: uncharacterized protein LOC105639235 [Jatropha curcas] |
| 10 |
Hb_005048_140 |
0.2076103443 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_011344_160 |
0.2079051017 |
- |
- |
PREDICTED: primary amine oxidase [Jatropha curcas] |
| 12 |
Hb_000172_540 |
0.2094865688 |
- |
- |
hypothetical protein POPTR_0005s08350g [Populus trichocarpa] |
| 13 |
Hb_000139_230 |
0.2103945852 |
transcription factor |
TF Family: SNF2 |
PREDICTED: probable ATP-dependent DNA helicase CHR12 [Jatropha curcas] |
| 14 |
Hb_013358_080 |
0.2108062192 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 25-like [Jatropha curcas] |
| 15 |
Hb_074197_020 |
0.2140701637 |
- |
- |
PREDICTED: uncharacterized protein LOC105645948 [Jatropha curcas] |
| 16 |
Hb_003998_010 |
0.2141359934 |
- |
- |
PREDICTED: protein BRASSINOSTEROID INSENSITIVE 1 [Jatropha curcas] |
| 17 |
Hb_000157_200 |
0.2146039786 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 isoform X3 [Eucalyptus grandis] |
| 18 |
Hb_000203_230 |
0.214899793 |
- |
- |
PREDICTED: probable inactive dual specificity protein phosphatase-like At4g18593 [Jatropha curcas] |
| 19 |
Hb_000815_130 |
0.2152349948 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 20 |
Hb_029904_020 |
0.217649874 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |