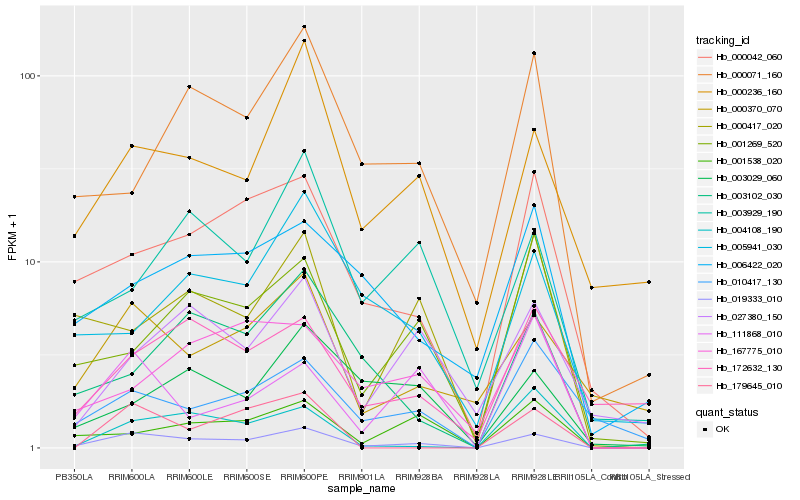
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019333_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633210 [Jatropha curcas] |
| 2 |
Hb_000042_060 |
0.2110153509 |
- |
- |
PREDICTED: uncharacterized protein LOC105632794 [Jatropha curcas] |
| 3 |
Hb_000370_070 |
0.2202842008 |
- |
- |
PREDICTED: subtilisin-like protease-like [Citrus sinensis] |
| 4 |
Hb_000236_160 |
0.220515179 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 6 [Jatropha curcas] |
| 5 |
Hb_000417_020 |
0.2312308585 |
- |
- |
PREDICTED: uncharacterized protein LOC105649674 [Jatropha curcas] |
| 6 |
Hb_001538_020 |
0.2366528923 |
- |
- |
PREDICTED: uncharacterized protein LOC105629608 [Jatropha curcas] |
| 7 |
Hb_001269_520 |
0.2378389991 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 8 |
Hb_004108_190 |
0.2386140312 |
- |
- |
PREDICTED: protein catecholamines up-like [Jatropha curcas] |
| 9 |
Hb_003102_030 |
0.2436032872 |
- |
- |
PREDICTED: uncharacterized protein LOC105644418 [Jatropha curcas] |
| 10 |
Hb_000071_160 |
0.2464510242 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 11 |
Hb_010417_130 |
0.2481960733 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 2 [Jatropha curcas] |
| 12 |
Hb_003929_190 |
0.2482135298 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 13 |
Hb_027380_150 |
0.2509014451 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_111868_010 |
0.2533250581 |
- |
- |
hypothetical protein CICLE_v10029531mg [Citrus clementina] |
| 15 |
Hb_172632_130 |
0.2537959447 |
- |
- |
hypothetical protein JCGZ_22584 [Jatropha curcas] |
| 16 |
Hb_006422_020 |
0.2546671525 |
- |
- |
PREDICTED: uncharacterized protein LOC105649338 [Jatropha curcas] |
| 17 |
Hb_167775_010 |
0.2566288418 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 18 |
Hb_179645_010 |
0.256757202 |
- |
- |
- |
| 19 |
Hb_005941_030 |
0.2587876283 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_003029_060 |
0.2593128443 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |