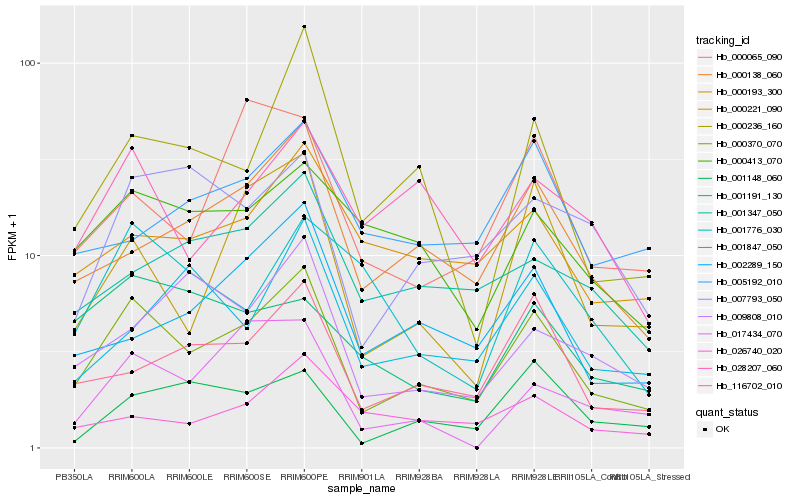
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000370_070 |
0.0 |
- |
- |
PREDICTED: subtilisin-like protease-like [Citrus sinensis] |
| 2 |
Hb_000138_060 |
0.1730430847 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 3 |
Hb_116702_010 |
0.1770413592 |
- |
- |
Leucine-rich repeat protein kinase family protein [Theobroma cacao] |
| 4 |
Hb_000221_090 |
0.1791586838 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-6-like [Jatropha curcas] |
| 5 |
Hb_000236_160 |
0.1859623939 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 6 [Jatropha curcas] |
| 6 |
Hb_028207_060 |
0.1872085221 |
- |
- |
PREDICTED: uncharacterized protein LOC105639235 [Jatropha curcas] |
| 7 |
Hb_007793_050 |
0.1876237526 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000193_300 |
0.1917410163 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |
| 9 |
Hb_000413_070 |
0.191980711 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002289_150 |
0.1963411085 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 11 |
Hb_001347_050 |
0.1980086744 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 12 |
Hb_001148_060 |
0.1992512852 |
- |
- |
hypothetical protein JCGZ_00278 [Jatropha curcas] |
| 13 |
Hb_026740_020 |
0.2059581897 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 14 |
Hb_017434_070 |
0.2084747163 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_009808_010 |
0.2088233049 |
- |
- |
Leucine-rich repeat protein kinase family protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_000065_090 |
0.209136589 |
- |
- |
potassium channel tetramerization domain-containing protein, putative [Ricinus communis] |
| 17 |
Hb_001847_050 |
0.2107723242 |
- |
- |
hypothetical protein CISIN_1g019843mg [Citrus sinensis] |
| 18 |
Hb_001776_030 |
0.2121431322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005192_010 |
0.2128436288 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 20 |
Hb_001191_130 |
0.2138361506 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g71810, chloroplastic [Jatropha curcas] |