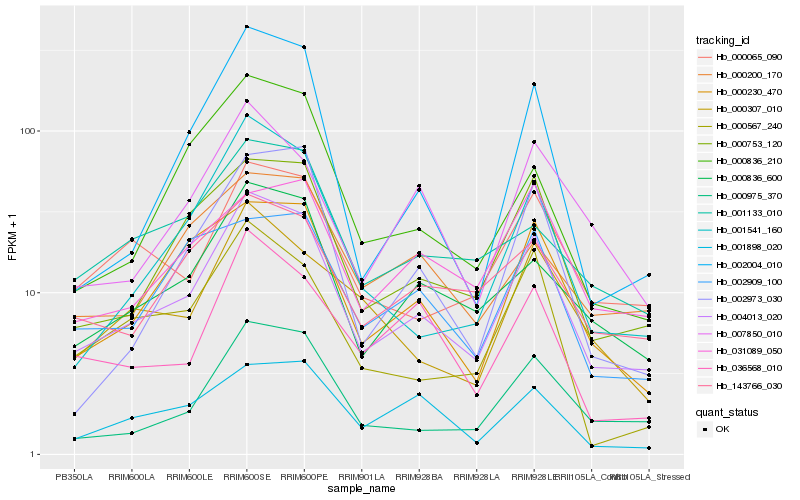
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004013_020 |
0.0 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 2 |
Hb_000753_120 |
0.107690612 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 3 |
Hb_000975_370 |
0.116074633 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB59 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000836_600 |
0.1181985486 |
- |
- |
PREDICTED: delta(8)-fatty-acid desaturase 2 [Jatropha curcas] |
| 5 |
Hb_000230_470 |
0.1266240648 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000065_090 |
0.1321607204 |
- |
- |
potassium channel tetramerization domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_001541_160 |
0.1337613704 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 8 |
Hb_143766_030 |
0.1473540327 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_000567_240 |
0.148305397 |
- |
- |
PREDICTED: F-box/LRR-repeat MAX2 homolog A-like [Jatropha curcas] |
| 10 |
Hb_001133_010 |
0.1486889628 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000836_210 |
0.1520738098 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 12 |
Hb_002004_010 |
0.1528516337 |
- |
- |
CBL-interacting protein kinase 9 [Populus trichocarpa] |
| 13 |
Hb_036568_010 |
0.1534284259 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 23 [Jatropha curcas] |
| 14 |
Hb_000307_010 |
0.1538826893 |
- |
- |
PREDICTED: ganglioside-induced differentiation-associated protein 2-like [Jatropha curcas] |
| 15 |
Hb_002973_030 |
0.1542345216 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 16 |
Hb_007850_010 |
0.1545852539 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 17 |
Hb_001898_020 |
0.154805708 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 18 |
Hb_031089_050 |
0.1550372315 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_000200_170 |
0.1558471273 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Populus euphratica] |
| 20 |
Hb_002909_100 |
0.1582131679 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |